Fig. 1. Cryo-EM structure of PRC2-AEBP2-JARID2 bound to a H2AK119ub1 nucleosome.
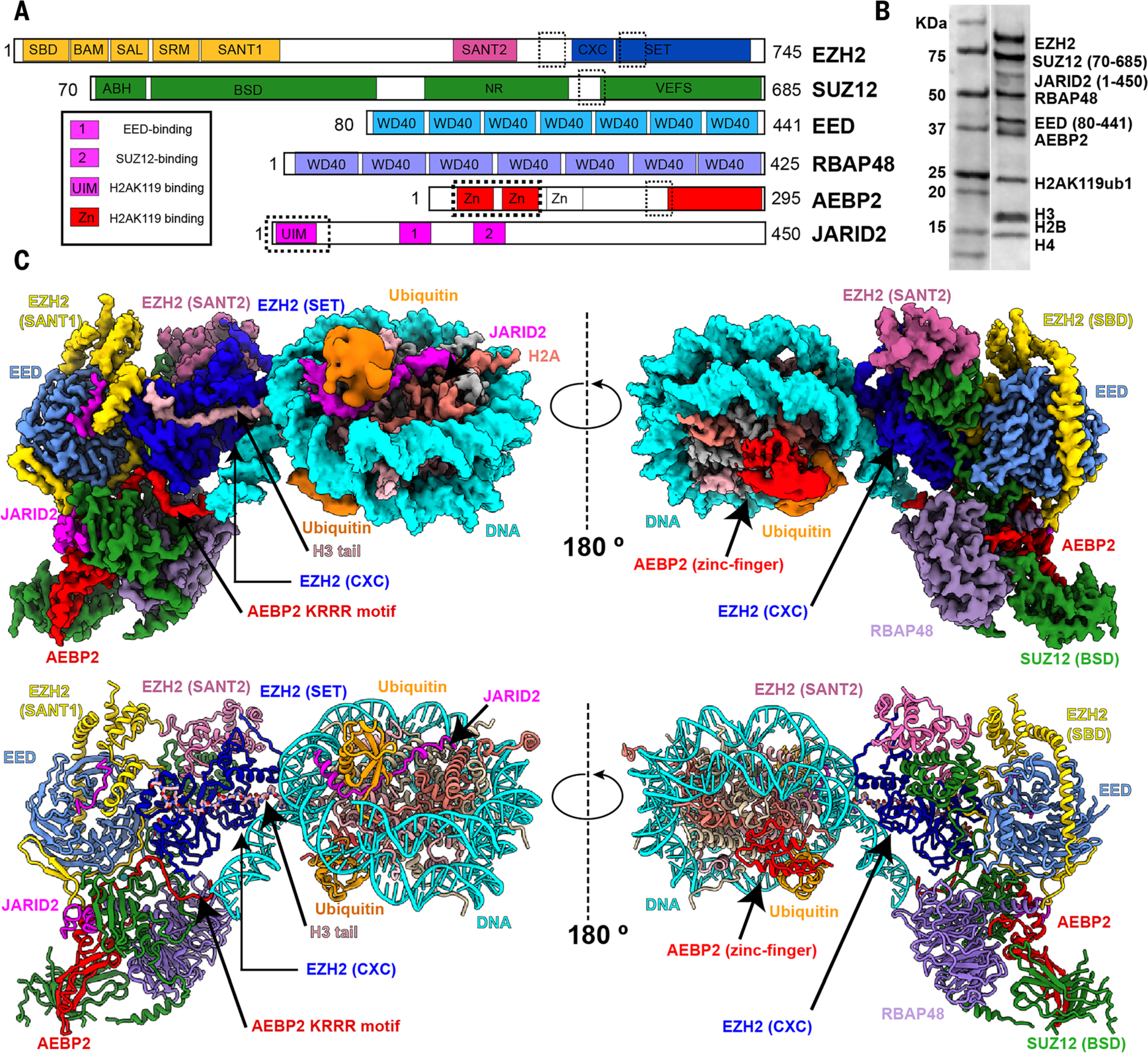
(A) Schematic representation of the proteins in the PRC2-AEBP2-JARID2 complex. Newly modeled regions in EZH2, JARID2, and AEBP2 that contribute to the interaction with the nucleosome are marked by dashed boxes. (B) Coomassie-stained gel showing all of the PRC2-core subunits, cofactors JARID2 and AEBP2, and histone proteins in the sample used for structural studies. (C) (Top) Cryo-EM density map for PRC2-AJ1–450 bound to an H2AK119ub1-containing nucleosome. (Bottom) Atomic model of PRC2-AJ1–450 bound to an H2AK119ub1-containing nucleosome, with EZH2 (SANT1) highlighted in gold, EZH2 (SANT2) in hot pink, EZH2 (SET) in blue, EED in light blue, RBAP48 in light purple, SUZ12 in green, JARID2 in magenta, AEBP2 in red, ubiquitin in orange, histone H3 in light pink, H2A in salmon, H4 and H2B in khaki, and nucleosome DNA in cyan. The image shown is a composite map where the PRC2 and nucleosome core correspond to the local resolution-filtered map from multibody refinement, whereas the ubiquitin, JARID2 UIM, and AEBP2 zinc finger densities correspond to the local resolution-filtered map from the consensus refinement. This color coding also applies to movie S1.
