Fig. 2. Interaction of EZH2 with the histone H3 tail and nucleosomal DNA.
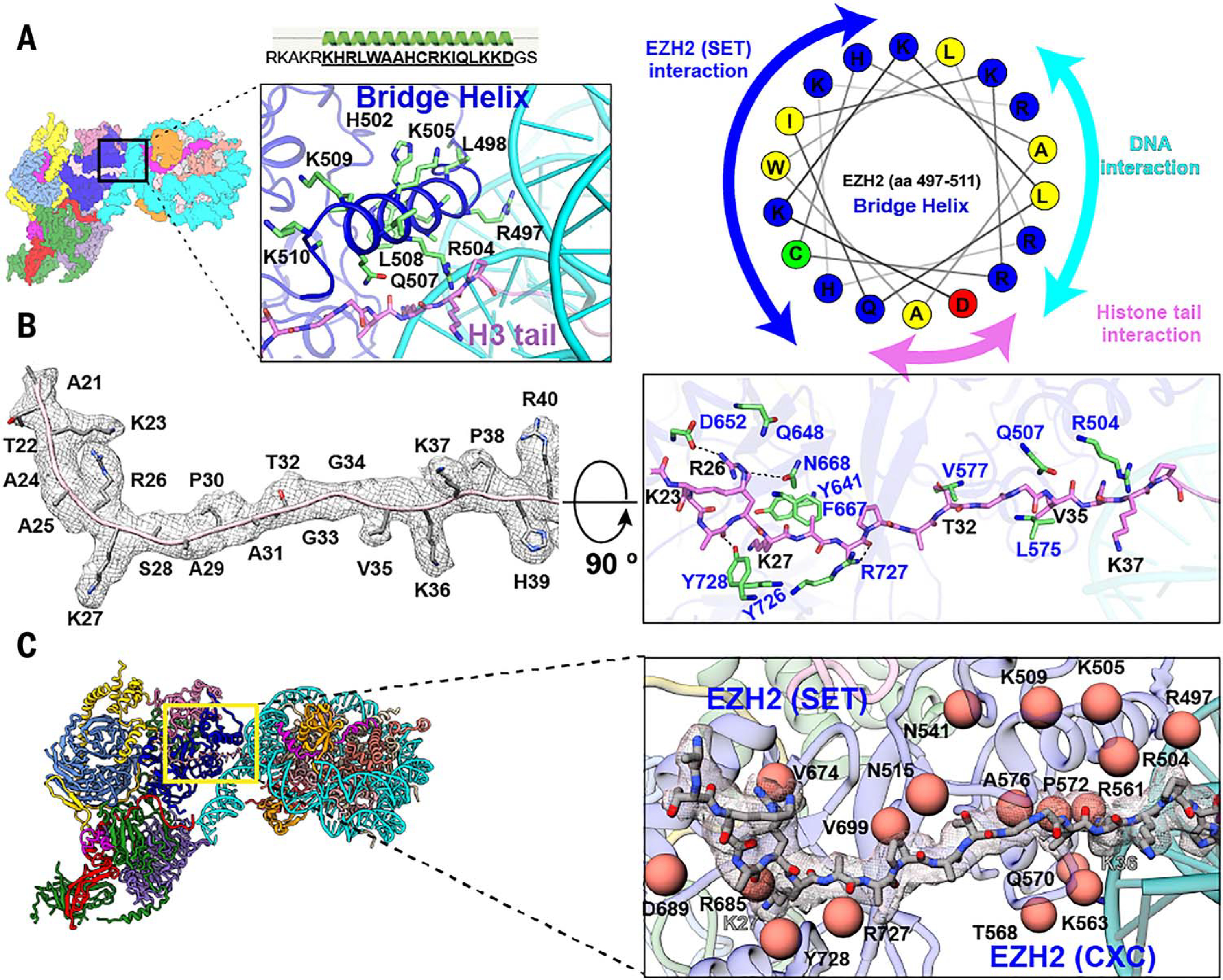
(A) (Top) Cartoon representation of the newly defined EZH2 bridge helix (amino acids 497 to 513) that interacts with nucleosomal DNA and the H3 tail. (Left) Cryo-EM structure of the PRC2-AEBP2-JARID2 complex bound to an H2AK119ub1-containing nucleosome. The zoom out shows the bridge helix, with residues interacting with the EZH2 (SET) domain, nucleosomal DNA, and the histone tail depicted in stick representation. (Right) Helix wheel diagram for the bridge helix that shows the distribution of positively charged residues on the nucleosomal DNA interacting face (cyan), positive and hydrophobic residues on the EZH2 (SET) interacting face (blue), and the residues interacting with the backbone of the H3 tail (pink). aa, amino acids. (B) (Left) Density map of the histone H3 tail (amino acids 21 to 40) (contour level: 0.024) with the corresponding atomic model. Clear density for residues R26 and K27, which have been previously observed, as well as density for K23, K36, K37, and V35 allow us to define the full extent of interaction between EZH2 (SET) and the histone H3 tail. (Right) Extensive electrostatic and van der Waals interactions between the residues in EZH2 (SET) (blue; shown in green stick representation) and the histone H3 tail (black; shown in pink stick representation) guide the H3 tail into the catalytic site. (C) Close-up view of the interaction of EZH2 (SET and CXC) with the histone H3 tail (stick representation with corresponding cryo-EM density in transparency), which shows residues involved in either direct or indirect interaction with the histone H3 tail and nucleosome DNA that are also frequently found mutated in cancers as orange spheres [COSMIC database (57)]. Single-letter abbreviations for the amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; and Y, Tyr.
