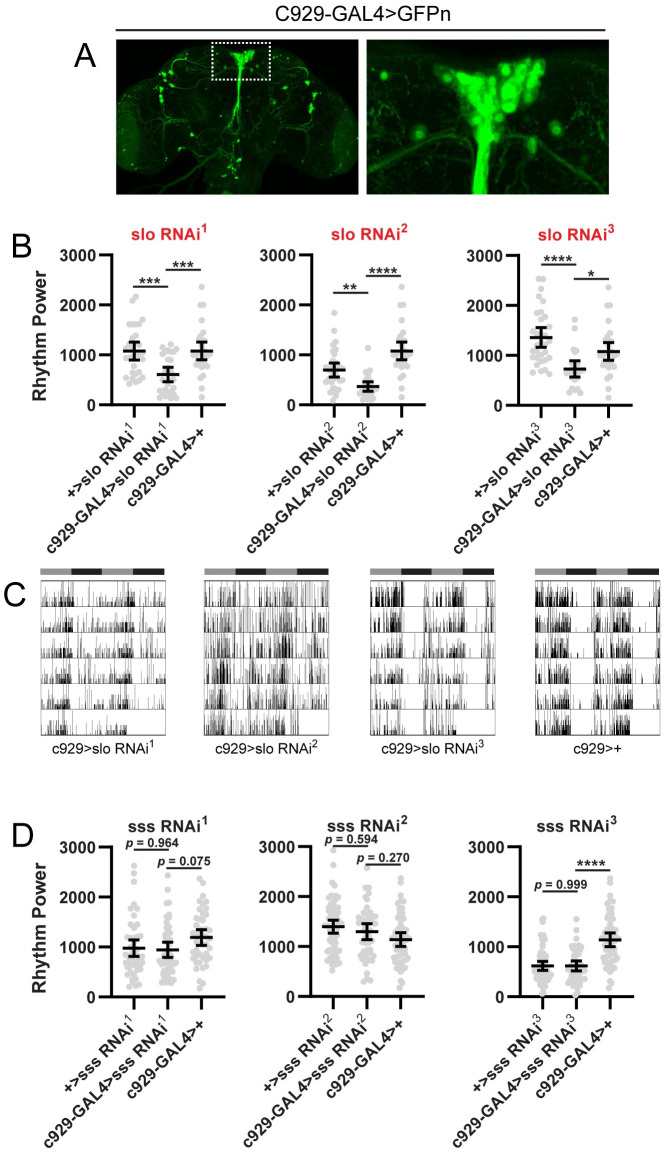
Fig 5. Confirmation of slo as a circadian output gene.
(A) Representative maximum projection confocal images of a fly brain in which C929-GAL4 was used to drive expression of a nuclear-localized GFP (C929-GAL4>GFPn). GFP is expressed in all PI cells plus a number of peptidergic neurons outside the PI. The right panel shows a magnified view of the boxed region on the left panel. (B) Rest:activity rhythm power is displayed for flies in which C929-GAL4 was used to drive expression of 3 independent RNAi constructs targeting the slo gene. Lines are means ± 95% confidence intervals. Dots represent individual flies. *p <0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001, Tukey’s multiple comparisons test for experimental line compared to GAL4 or UAS controls. (C) Representative single fly activity records over 6 days in DD for the genotypes listed. (D) Rest:activity rhythm power is displayed as described in (A) for flies in which C929-GAL4 was used to drive 3 independent RNAi constructs targeting the sss gene. Graphs labeled in red indicate experiments for which we observed significantly reduced rest:activity rhythm strength in experimental flies compared to both GAL4 and UAS controls. C929-GAL4-mediated expression of all 3 slo-targeting RNAi constructs significantly reduced rest:activity rhythm strength, but expression of sss-targeting RNAi constructs was without effect. See S2 File for detailed information on rest:activity rhythm power, period and n for each line.