Figure 6. FBXO32 is necessary to restrain HrasG12V tumor initiation and growth.
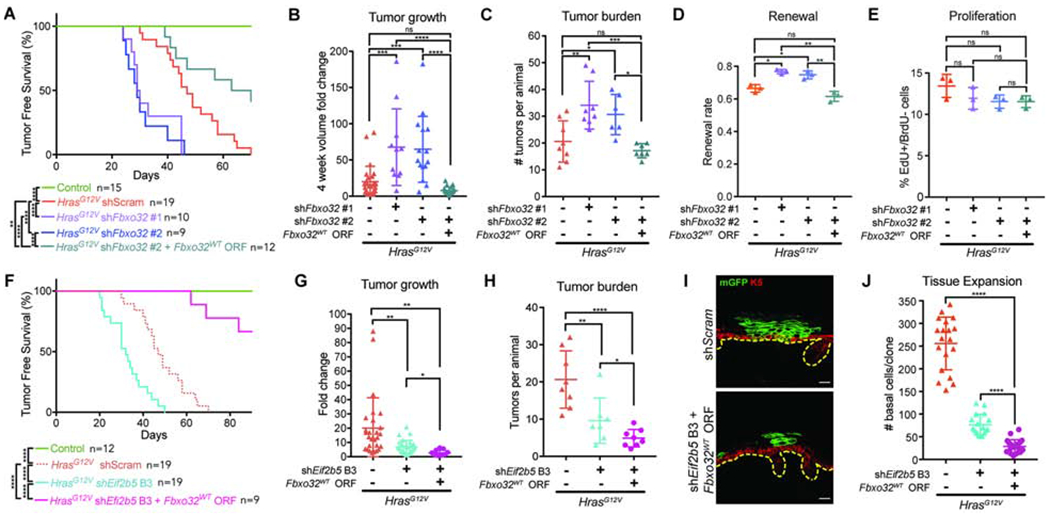
A) Tumor-free survival of WT (control) and transduced HrasG12V animals. Log-rank test used for significance analyses.
B) Tumor volume fold change 4 weeks after tumor initiation. n=27 HrasG12V+shScram, 11 HrasG12V+shFbxo32 #1, 15 HrasG12V+shFbxo32 #2, and 21 HrasG12V+shFbxo32 #2+Fbxo32WT tumors. 4 animals were assessed per condition.
C) Number of tumors per animal within 4 weeks of first tumor initiation. n=8 HrasG12V+shScram, 8 HrasG12V+shFbxo32 #1, 6 HrasG12V+shFbxo32 #2, and 7 HrasG12V+shFbxo32 #2+Fbxo32WT animals.
D-E) (D) Renewal probability and (E) proliferation rate in tumors of transduced HrasG12V animals. n=3 animals per condition. Approx. 100 cell divisions (for renewal assay) or 100 basal cells (for proliferation assay) were scored per animal.
F) Tumor-free survival of WT (control) or transduced HrasG12V animals. Log-rank test used for significance analyses.
G) Tumor volume fold change 4 weeks after tumor initiation. n=27 HrasG12V+shScram, 27 HrasG12V+shEif2b5 B3, and 15 HrasG12V+shEif2b5 B3+Fbxo32WT tumors. 4 animals were assessed per condition.
H) Number of tumors per animal within 4 weeks of first tumor initiation. n=8 HrasG12V+shScram, 7 HrasG12V+shEif2b5 B3, and 8 HrasG12V+shEif2b5 B3+Fbxo32WT animals.
I) Representative cross-section of transduced HrasG12V clone. mGFP (green) marks transduced epidermal cells, K5 (red) marks basal progenitors, and dashed line marks basement membrane. Scale bars, 25μm.
J) Basal cell numbers per clone in transduced E18.5 epidermis. n=18 HrasG12V+shScram, 17 HrasG12V+shEif2b5 B3, and 21 HrasG12V+shEif2b5 B3+Fbxo32WT clones. 3 animals were assessed per condition.
Mean and SD are shown. ANOVA with Tukey’s range test for multiple comparisons used to determine statistical differences. *:P-value<0.05; **:P-value<0.01; ***:P-value<0.001; ****:P-value<0.0001; ns=not significant. See also Figure S5.
