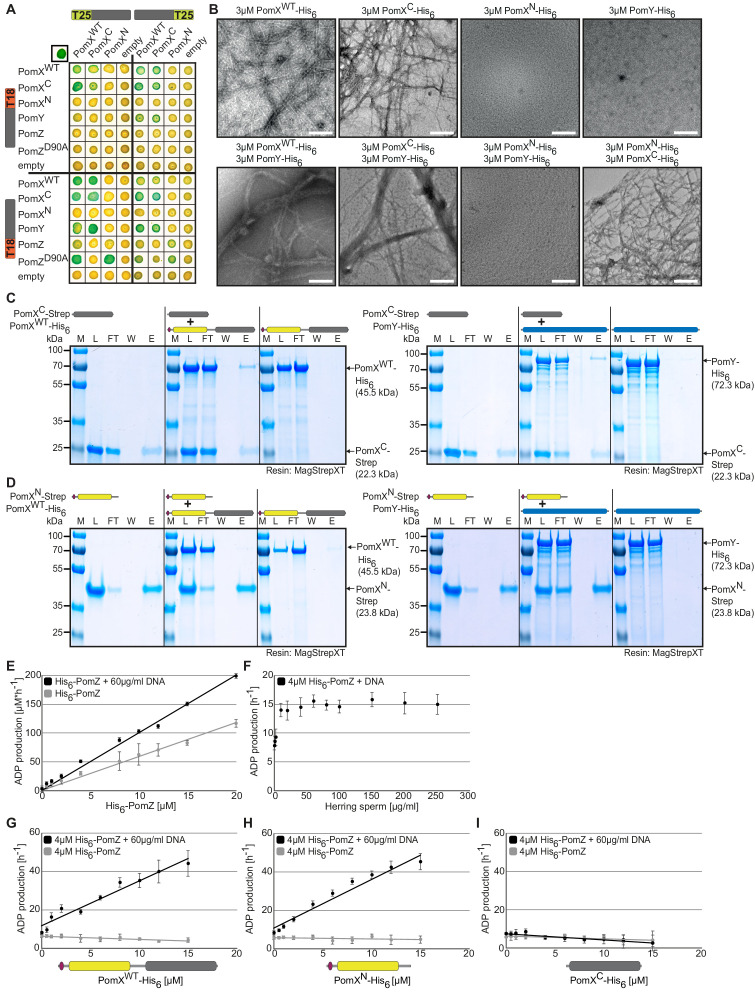
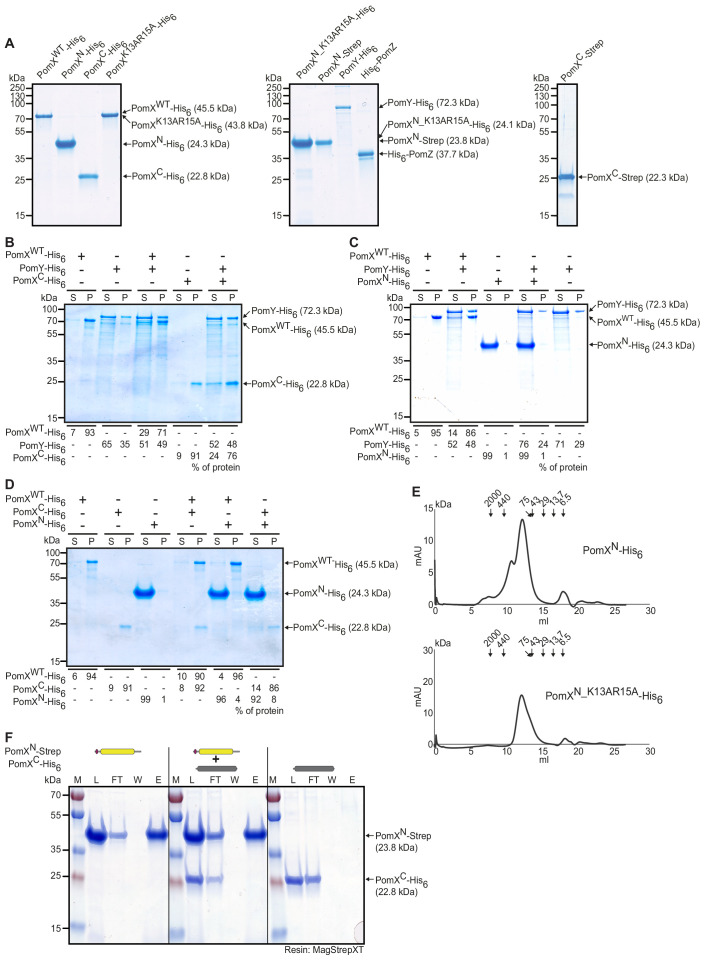
Figure 2. PomXC interacts with PomX and PomY while PomXN stimulates PomZ ATPase activity.
(A) BACTH analysis of interactions between Pom proteins. The indicated protein fragments were fused to T18 and T25 as indicated. Blue colony indicates an interaction, white no interaction. Positive control in upper left corner, leucine zipper of GCN4 fused to T25 and T18. For negative controls, co-transformations with empty plasmids were performed. Images show representative results and were performed in three independent experiments. (B) TEM images of negatively stained purified proteins. Proteins were applied to the EM grids alone or after mixing in a 1:1 molar ratio as indicated before staining. Scale bar, 200 nm. Images show representative results of several independent experiments. (C, D) In vitro pull-down experiments with purified PomXC-Strep, PomXN-Strep, PomXWT-His6, and PomY-His6. Instant Blue-stained SDS-PAGE shows load (L), flow-through (FL), wash (W), and elution (E) fractions using MagStrep XT beads in pull-down experiments with 10 µM of indicated proteins alone or pre-mixed as indicated on top. Molecular size markers are shown on the left and proteins analyzed on the right together with their calculated MW. Note that PomXWT-His6 (Schumacher et al., 2017) and PomXN-Strep migrate aberrantly and according to a higher MW. All samples in a panel were analyzed on the same gel and black lines are included for clarity. Experiments were repeated in two independent experiments with similar results. (E–I) His6-PomZ ATPase activity. ADP production rate was determined in an NADH-coupled photometric microplate assay in the presence of 1 mM ATP at 32°C. DNA and PomX variants were added as indicated. Spontaneous ATP hydrolysis and NADH consumption was accounted for by subtracting the measurements in the absence of His6-PomZ. Data points show the mean±STDEV calculated from six independent measurements.