(
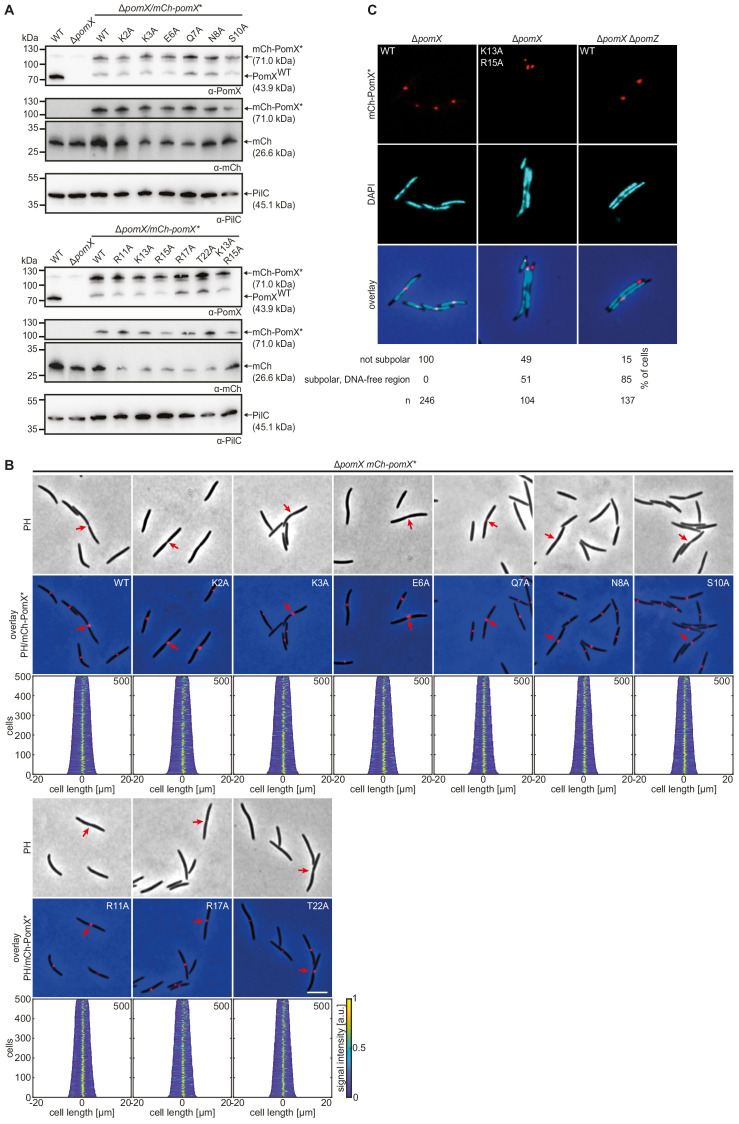
A) Western blot analysis of the accumulation of mCh-PomX variants in indicated strains. Protein from the same number of cells was loaded per lane. Molecular mass marker is shown on the left and analyzed proteins on the right. The same blots were sequentially analyzed with α-PomX (top panel), α-mCh (middle panel), and α-PilC (lower panel) antibodies. PilC was used as a loading control. Note PomX (43.9 kDa) does not migrate at the expected size in SDS-PAGE but instead as a protein of a molecular weight of 72 kDa. Similarly, mCh-PomX migrates at ~110 kDa. Similar results were obtained in two independent experiments. (
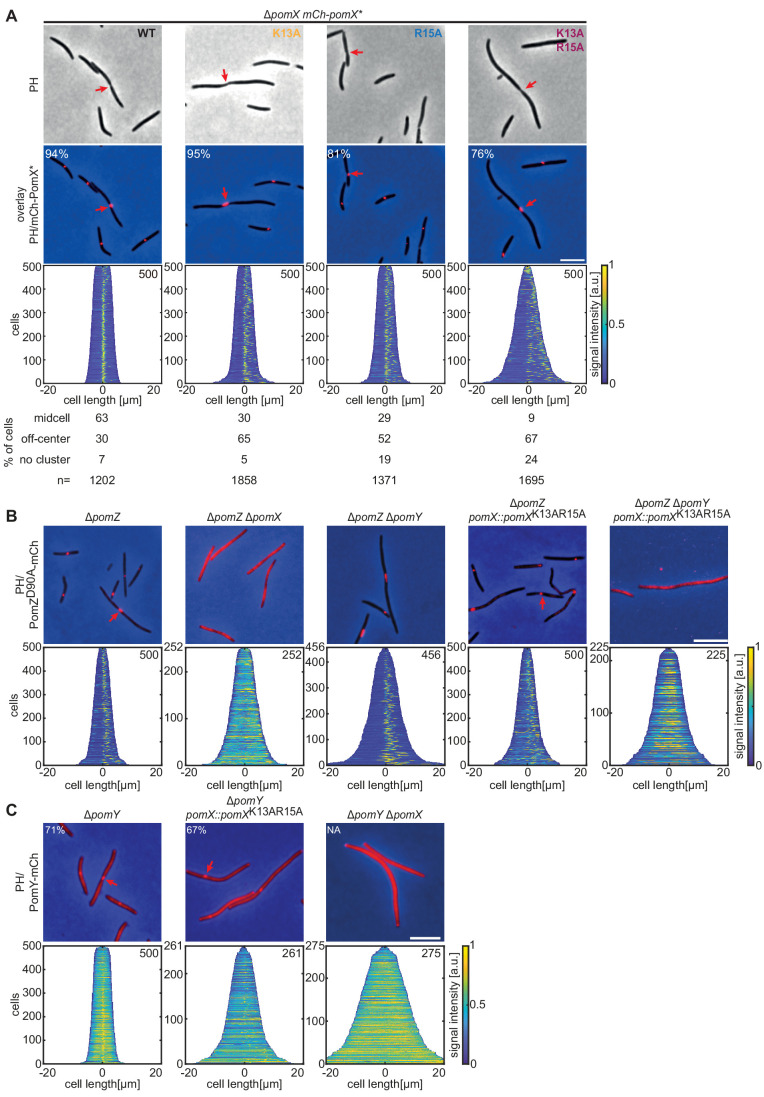
B) Fluorescence microscopy of indicated mCh-PomX variants. Phase-contrast and fluorescence images of representative cells and the overlay are shown. Red arrows indicate cell division constrictions. Scale bar, 5 µm. Demographs were created as in
Figure 1E. Experiments were repeated in two independent experiments with similar results. (
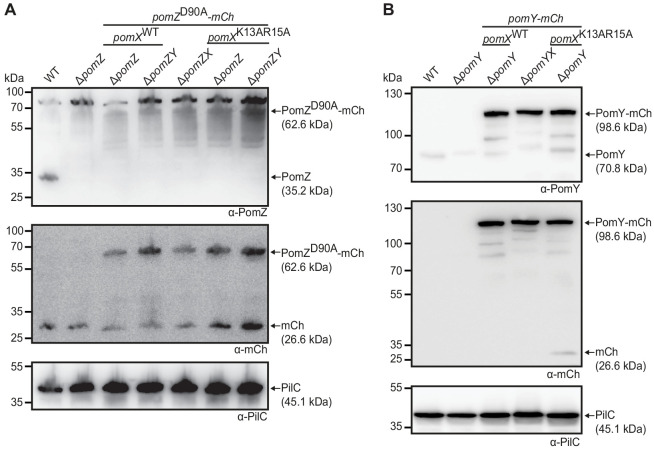
C) Fluorescence microscopy of mCh-PomX variants in DAPI-stained cells of the indicated genotype. The mCh signal (first panel), DAPI signal (second panel), and the overlay (third panel) show representative cells. Amino acid substitutions are indicated in white in the mCh images. Scale bar, 5 µm. Quantification of mCh-PomX* localization patterns in % and the number of analyzed cells is shown below the images. Images show representative cells. Similar results were obtained in two independent experiments.