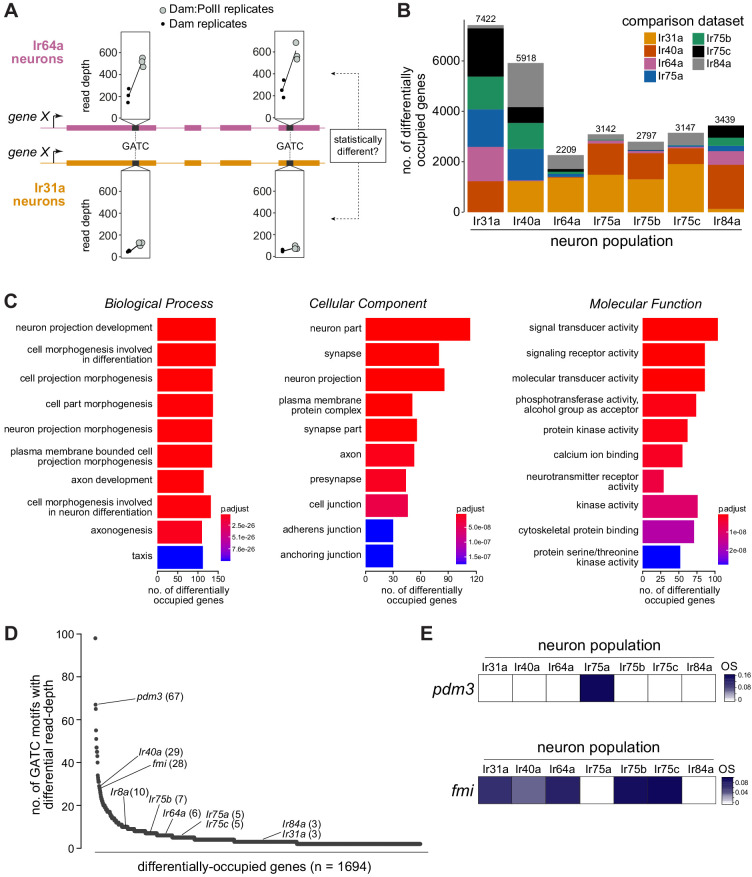
Figure 4. Differentially occupied genes across Ir populations.
(A) Schematic illustrating the identification of a differentially occupied gene between two Ir populations by using variation in read depth at GATC motifs (see Materials and methods). In this example, occupancy of ‘gene X’ is compared between Ir64a neurons and Ir31a neurons. At GATC motifs (black bars), a test is applied to determine if significant variation exists in the relative read depths (in triplicate Dam:PolII versus Dam-alone experiments) between these two neuron populations. Fictive data for two GATC motifs are depicted: for both, the relative read depth in the Dam:PolII experiments (compared to the Dam-alone experiments) is greater in the Ir64a neurons. (B) Stacked bar plots of the numbers of genes in the genome that are differentially occupied among Ir neuron populations based on pairwise tests (see Materials and methods). The colors indicate the proportions of genes emerging from comparisons with the six other neuron populations. The figures above the bars are the total numbers of genes from all pairwise comparisons; note that a given gene may be counted multiple times. (C) Gene Ontology (GO) analysis of the 1694 differentially occupied genes, showing the top ten over-represented GO terms. The x-axis represents the number of genes annotated to a particular GO term in the input subset. The value indicated by the colored bar is the probability of observing at least the same number of genes associated to that GO term compared to what would have occurred by chance. (D) Ranking of the 1694 differentially occupied genes ordered by the number of GATC motifs contributing to their between-neuron differences (uncorrected for gene length). (E) OS heatmaps (calculated as in Figures 1–2) of pdm3 and fmi in the seven Ir neuron populations.