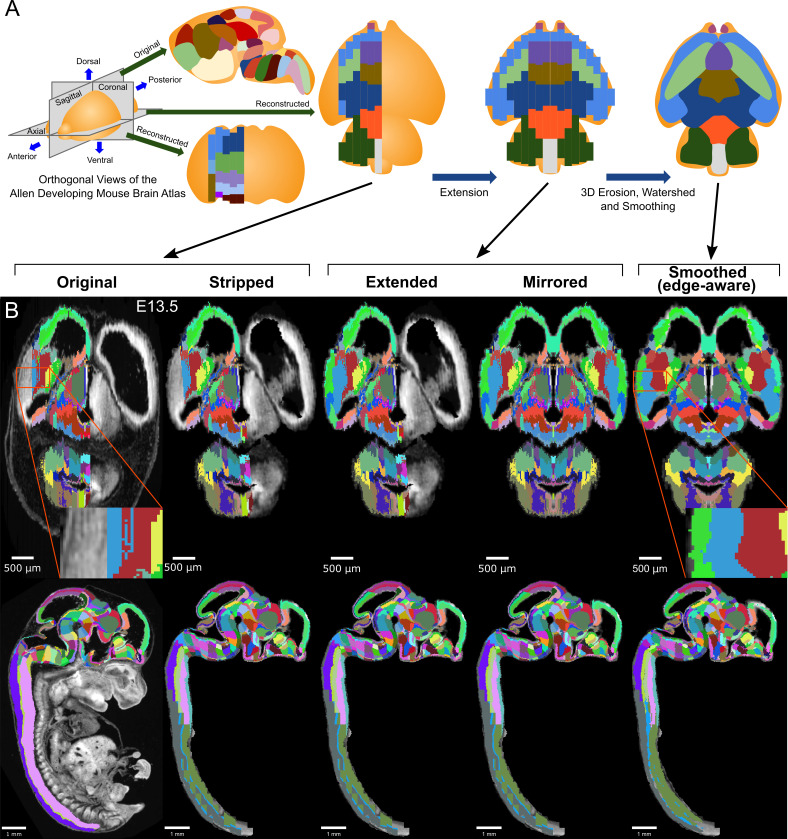
Figure 1. 3D atlas refinement pipeline overview and full example in the E13.5 atlas.
(A) 2D-derived atlases, such as those in the Allen Developing Mouse Brain Atlas, are smooth and consistent in the sagittal plane in which they were annotated. However, in the 3D reconstructions of these 2D sagittal planes, the coronal and axial planes reveal missing sections and jagged edges. To improve their performance for annotating 3D data, the lateral edges are extended to complete the labeled hemisphere. A 3D rotation is applied to bring the brain parallel to the image borders, then both the completed hemisphere labels and underlying microscopy sections are mirrored across the sagittal midline to complete coverage in the opposite hemisphere. To improve anatomical registration, the labels are each eroded and re-grown through a 3D watershed, guided by the anatomical edge map. To smooth the final product, labels are iteratively filtered with a morphological opening operation, or a closing operation for small, disconnected labels that would otherwise be lost. (B) The pipeline illustrated in axial (top) and sagittal (bottom) views on the ADMBA E13.5 atlas, which requires the full pipeline shown in ‘A’, including an additional step to strip out non-CNS tissue from the original whole-embryo imaging. The nomenclature for pipeline steps shown here is used consistently throughout the manuscript. Insets of the ‘Original’ and ‘Smoothed (edge-aware)’ lateral regions highlight the label extension and smoothing. A spreadsheet mapping colors to label names and IDs in the Allen ontology for each atlas in this manuscript can be found in Supplementary file 1.