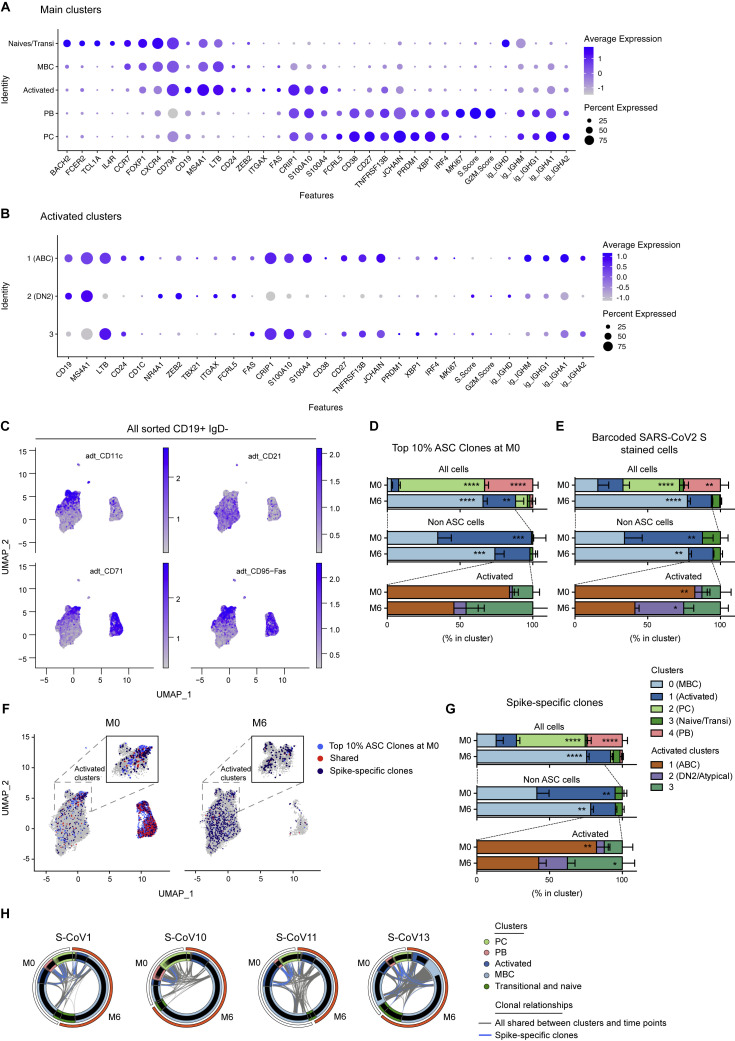
Figure S2.
Gene expression markers and cluster assignments for the main single-cell populations identified in acute and convalescent S-CoV patients, Related to Figure 2
(A and B) Dot plots showing expression of selected genes in cells from the main clusters (A) or in cells from the “Activated” cluster (B). Size of dots represents the percentage of cells in the cluster in which transcripts for that gene are detected. Dot color represents the average expression level (scaled normalized counts) of that gene in the population.
(C) Feature plots showing scaled normalized counts for CD21, CD71, CD11c, CD95 barcoded antibodies in all sorted CD19+IgD-.
(D and E) Relative cluster distribution at M0 and M6 for cells belonging to one of the top 10% ASC clones (D) or stained by the barcoded-anti-His/His-tagged SARS-CoV-2-S protein combination (barcoded-SARS-CoV-2-S) (E). Top panels represent cluster distribution for all cells, middle panels represent cluster distribution for non-ASC cells and bottom panels represent cluster distribution for cells belonging to the “Activated” cluster. Bars indicate mean with SEM.
(F) UMAP of all cells at M0 or M6, with cells belonging to one of the top 10% ASCs clones highlighted (light blue). Cells belonging to Spike-specific clones, as defined in the Methods section, are also highlighted (red when members of one of the top 10% ASC clones at M0, dark blue otherwise).
(G) Relative cluster distribution at M0 and M6 for cells belonging to spike-specific clones. Top panel represent cluster distribution for all cells, middle panel represent cluster distribution for non-ASC cells and bottom panels represent cluster distribution for cells belonging to the “Activated” cluster. Bars indicate mean with SEM.
(H) Circos plots showing clonal relationships between cells from different UMAP clusters and time points. Blue lines indicate clonal relationships with one of the spike-specific clones and gray line all other clonal relationships.
RM two-way ANOVA and Sidak’s multiple comparison tests (D-E, G). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.01, ∗∗p < 0.01, ∗p < 0.05.