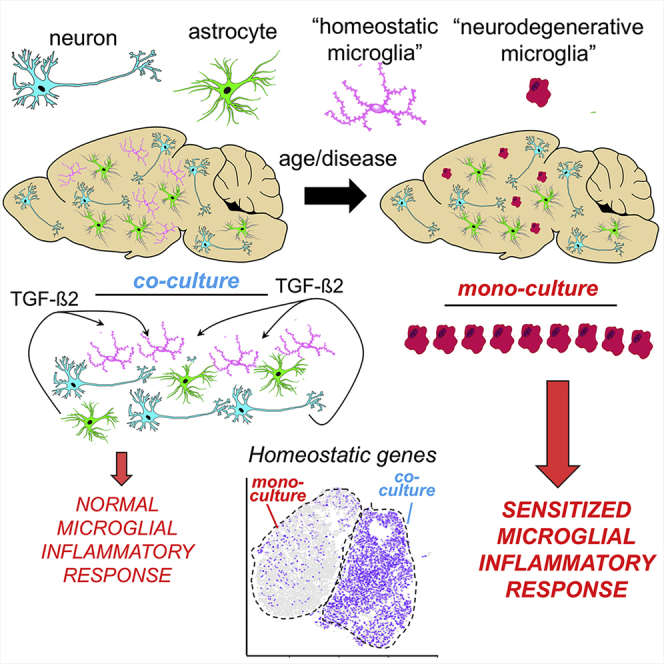
Summary
Microglia, brain-resident macrophages, require instruction from the CNS microenvironment to maintain their identity and morphology and regulate inflammatory responses, although what mediates this is unclear. Here, we show that neurons and astrocytes cooperate to promote microglial ramification, induce expression of microglial signature genes ordinarily lost in vitro and in age and disease in vivo, and repress infection- and injury-associated gene sets. The influence of neurons and astrocytes separately on microglia is weak, indicative of synergies between these cell types, which exert their effects via a mechanism involving transforming growth factor β2 (TGF-β2) signaling. Neurons and astrocytes also combine to provide immunomodulatory cues, repressing primed microglial responses to weak inflammatory stimuli (without affecting maximal responses) and consequently limiting the feedback effects of inflammation on the neurons and astrocytes themselves. These findings explain why microglia isolated ex vivo undergo de-differentiation and inflammatory deregulation and point to how disease- and age-associated changes may be counteracted.
Keywords: microglia, astrocytes, neurons, transcriptomics, ageing, neurodegeneration, RNA-seq, signal transduction
Graphical abstract
Highlights
-
•
Neurons and astrocytes combine to promote the microglial homeostatic signature
-
•
This rescues changes that happen in microglia ex vivo and in disease
-
•
Mechanistically, this involves transforming growth factor beta 2 (TBGF-β2) signaling
-
•
Neurons and astrocytes also repress microglial inflammatory responses
Baxter et al. show that the transformation of microglia from a healthy to disease-associated state can be suppressed by the combined actions of neurons and astrocytes via a mechanism involving TBGF-β2 signaling. They also repress exaggerated microglial responses to mild stimuli and limit the feedback signaling from activated microglia.
Introduction
Arising from yolk sac erythromyeloid progenitors, microglia are brain-resident macrophages that play critical roles in normal brain development and maturity (Wu et al., 2015). Recent evidence suggests that dysregulation of microglia function contributes to or drives the pathogenesis of certain neurodevelopmental and psychiatric diseases such as autism and schizophrenia, as well as neurodegenerative diseases, including Alzheimer’s disease (AD) (Perry and Holmes, 2014; Ransohoff, 2016; Salter and Stevens, 2017). Thus, identifying the mechanisms that control microglia function, including the impact of genetic and environmental influences, is important for understanding normal brain function and designing therapeutic strategies for disease intervention.
During development, microglia acquire a gene expression signature that distinguishes them from other macrophage populations and adopt a ramified morphology important for their role in immune surveillance (Butovsky et al., 2014; Matcovitch-Natan et al., 2016; Nimmerjahn et al., 2005). However, maintenance of these characteristics requires continued instruction from the CNS microenvironment. When maintained in culture, microglia generally assume a non-ramified, amoeboid morphology resembling microglia in injured tissues (Stansley et al., 2012). Moreover, mature microglia rapidly de-differentiate ex vivo, losing their signature gene expression profile (Bohlen et al., 2017; Gosselin et al., 2017). This signature can be re-acquired by engrafting the cells back into the brain (Bohlen et al., 2017), which is evidence that microglial identity requires continuous instruction from the CNS microenvironment. This environment contains multiple cell types, including neurons, macroglia, and vascular components, although which are important for defining microglial identity is unclear. Related to the control and specification of microglial homeostatic identity is their consequent response to inflammatory challenges. Signals that can influence microglial basal state are frequently implicated in immunomodulation. Signals derived from neurons and macroglia (particularly astrocytes) implicated in modulating microglial inflammatory responses include both secreted and cell surface ligands (Hoarau et al., 2011; Perry and Holmes, 2014).
Here, we investigated whether specific CNS cell types are sufficient to promote microglial identity. We hypothesized that neurons and/or astrocytes may be able to provide a surrogate CNS environment that promotes expression of mature microglial signature genes ordinarily lost when microglia are maintained in vitro, as well as physiologically relevant immunomodulatory cues. We initially used a recently described approach of mixed-species RNA sequencing (RNA-seq), a tool for elucidating non-cell-autonomous control of gene transcription (Qiu et al., 2018). Following the co-culture of purified neurons, astrocytes, and microglia from different species (mouse, human, and rat, respectively), individual cell-type transcriptomes can be profiled through the species-specific sorting of bulk RNA-seq data using the Python tool Sargasso (Qiu et al., 2018), avoiding gene expression artifacts and imperfections associated with physical sorting (Okaty et al., 2011a, 2011b; van den Brink et al., 2017; Wylot et al., 2015). We find that neurons and astrocytes can be employed in combination to secrete factors, including transforming growth factor β2 (TGF-β2), that uniformly promote microglial signature gene expression ordinarily lost upon isolation ex vivo (confirmed by single-cell RNA-seq) and lost in disease—the so-called microglial neurodegenerative phenotype (MGnD) (Krasemann et al., 2017). These signals also modulate microglial responses to inflammatory stimuli and, consequently, the impact of microglia on the neurons and astrocytes themselves. This platform, which maintains microglial identity and at least partly recapitulates the immunomodulatory signals that it receives in vivo from other CNS cell types, overcomes several issues with studying microglia in vitro, namely their de-differentiation and inflammatory deregulation, and enables the non-cell-autonomous consequences of microglial activation to be characterized.
Results
Neurons and astrocytes combine to drive microglial signature gene expression
We investigated, using our mixed-species RNA-seq approach (Qiu et al., 2018), whether neurons and astrocytes can promote expression of mature microglial signature genes ordinarily lost when microglia are maintained in vitro (Bohlen et al., 2017; Gosselin et al., 2017; Stansley et al., 2012). We followed our published protocol of co-culturing purified neurons and astrocytes from different species (mouse and human respectively), followed by seeding rat microglia either onto these co-cultures or on their own to create microglial monocultures. Other than the presence or absence of neurons and astrocytes, the microglia in co-culture or monoculture were treated the same and plated into the same defined, serum-free medium. Initial mixed-species RNA-seq characterization of the neuron-astrocyte co-culture confirmed that both cell types expressed the expected markers. The astrocyte-specific genes GFAP, GJA1, CLU, and VIM were all within the top 100 genes (by Fragments Per Kilobase of transcript per Million mapped reads [FPKM]) in the human astrocytes, and the neuron-specific genes Tubb3, Syp, Mapt, and Vamp2 were within the top 100 expressed genes in the mouse cortical neurons (Table S1, left and middle). Moreover, the astrocytes (but not neurons) were immuno-positive for GFAP (Figure S1A) and showed characteristic induction of a stellate morphology and expression of AQP4, SLC1A2, and SLC1A3 by neurons (Hasel et al., 2017).
To further profile the human astrocytes in our study, we utilized our genome-wide transcriptome data, comparing it to published data relating to acutely purified human astrocytes, neurons, microglia, and oligodendrocytes (Zhang et al., 2016). We calculated the ratio of expression of genes in acutely purified astrocytes relative to acutely purified neurons, microglia, and oligodendrocytes and plotted it against the ratio of expression of genes in the astrocytes used in our study relative to the same acutely purified neurons, microglia, and oligodendrocytes.
We observed a significant correlation in gene enrichment/de-enrichment in our astrocytes versus acutely purified astrocytes (relative to neurons, microglia, and oligodendrocytes; Figures S2A–S2C). We also curated the set of 100 genes most highly enriched in purified human astrocytes compared to neurons, microglia, and oligodendrocytes (Zhang et al., 2016) and observed that this gene set was also strongly expressed in the astrocytes used in our study relative to neurons, microglia, and oligodendrocytes (Figures S2D–S2F). Finally, we simply plotted the expression of genes in our astrocytes against that in acutely purified astrocytes and found a significant correlation (Figure S2G). Collectively these analyses provide evidence in favor of the astrocytic identity of the human astrocytes used in our study, although some differences from astrocytes acutely isolated from human brains cannot be ruled out.
Microglia were plated onto the neuron-astrocyte co-culture or maintained as a monoculture (Figures S1A and 1A). Cellular viability was measured by calculating (blind) levels of nuclear pyknosis (from DAPI-stained cultures), as in previous studies (Martel et al., 2012; Papadia et al., 2008), and viability 72 h after microglial platedown was good (89.6% ± 0.7% in monoculture [n = 3] and 86.7% ± 0.7% in co-culture [n = 3]). Moreover, in the co-culture the microglia distributed evenly across the plate, in proximity with neurons and astrocytes (Figure 1A). Mass spectrometry of extracellular medium in neuron-astrocyte co-cultures revealed extracellular matrix proteins (Table S1, right), including several collagens and matrix metalloproteinases and all members of the chondroitin sulfate proteoglycan lectican family (brevican, aggrecan, versican, and neurocan), which interact with hyaluronic acid in peri-neuronal nets in a manner stabilized by hyaluronan and proteoglycan link protein (also present). The presence of chondroitin sulfate proteoglycan brevican (Figure S2H) and N-acetylgalactosamine-beta-1-modified glycoproteins (Figure S2I), which are found in peri-neural nets (Slaker et al., 2016), was confirmed by immunohistochemistry. Thus, microglia in co-culture are exposed to neurons, astrocytes, and at least some elements of the brain’s extracellular matrix.
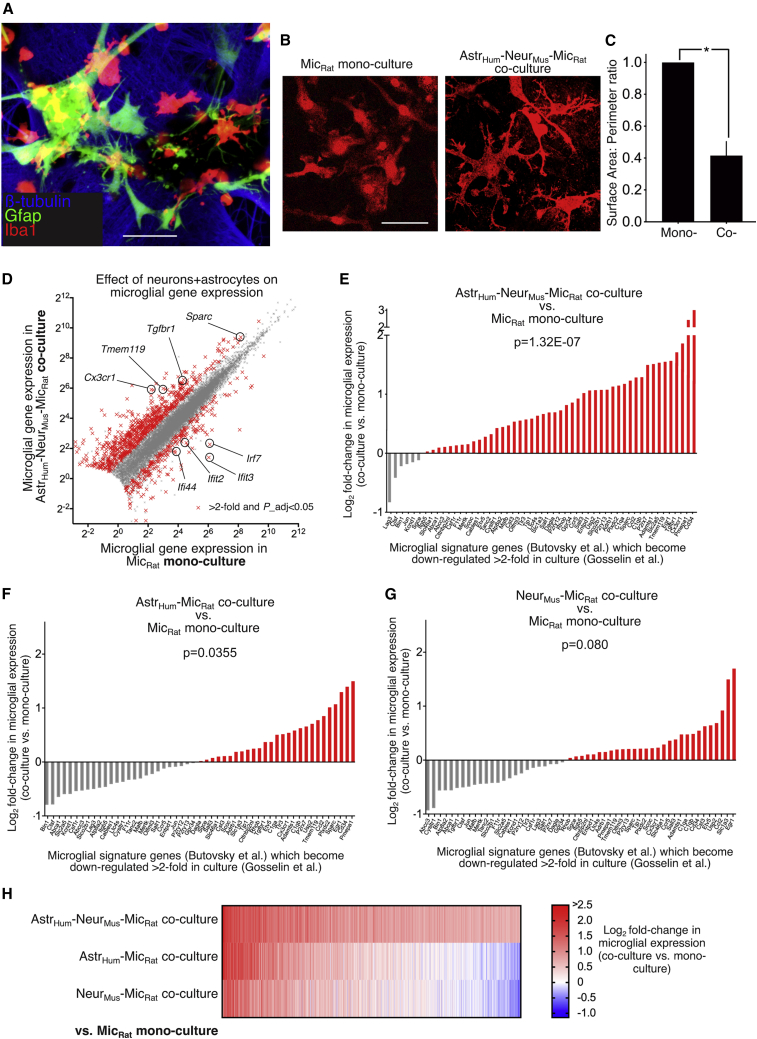
Figure 1.
Neurons and astrocytes combine to drive microglial homeostatic signature gene expression
(A) A triple stain of the astrocyte/neuron/microglia co-culture with the indicated antibodies. Scale bar, 50 μm.
(B) Example images of Iba1-stained microglia in mono- or co-cultures as indicated. Scale bar, 20 μm.
(C) Surface area/perimeter ratio was calculated in mono- and co-culture. ∗p = 0.007 (t = 6.662, degrees of freedom [df] = 3), paired t test (120 cells analyzed per condition across n = 4 independent biological replicates).
(D) The influence of neuron-astrocyte co-culture on the microglial transcriptome. RNA-seq was performed on RNA extracted from (rat) microglia mono-cultures and (rat) microglia co-cultured with (human) astrocytes and (mouse) neurons. Both sets of reads were subjected to the same Sargasso workflow to identify unambiguously rat (microglial) reads and FPKM of all 13,406 genes shown that average >1 FPKM across the datasets is plotted for mono-culture (x axis) versus co-culture (y axis). Highlighted with red crosses are the 982 genes whose expression is significantly changed (DESeq2 P_adj < 0.05) by >2-fold (n = 7 mono-culture; n = 4 co-culture). “N” refers here and throughout as independent biological replicates derived from different culture material on different occasions. Upregulated genes highlighted are example MHSGs (Butovsky et al., 2014); downregulated genes highlighted are example interferon-related gene (IRG) cluster members (Friedman et al., 2018).
(E) Neurons and astrocytes combined boost microglial signature genes which become suppressed in vitro. Genes considered are those expressed >0.5 FPKM in our data and within the group of microglia signature genes defined by Butovsky et al. (2014), which are downregulated >2-fold after microglia were maintained in culture for 7 days compared to their expression immediately post-isolation from the intact brain, according to the data of Gosselin et al. (2017). For each gene, log2 fold change (log2FC) in microglial gene expression is shown relative to microglial mono-culture in (rat) microglia (mouse) neuron (human) astrocyte co-culture. The data were mined from the complete set shown in (D). ∗p = 1.3E-07, F (1,477) = 28.69 relates to main effect of co- versus monoculture condition on the gene set, two-way ANOVA.
(F and G) RNA-seq was performed on RNA extracted from (rat) microglia monocultures and (rat) microglia co-cultured with (human) astrocytes (F) or (mouse) neurons alone (G). log2FC in microglial gene expression is shown relative to microglial monoculture of the same genes as in (E). p = 0.036, F(1,424) = 4.45 (F), p = 0.080, F(1,424) = 3.09 (G), two-way ANOVA.
(H) Heatmap of the log2FC in microglial gene expression in the three different types of co-culture. 1,550 genes significantly induced >1.5-fold (DESeq2 P_adj < 0.05) are shown.
Consistent with previous studies, we observed that monocultures of isolated microglia exhibited an amoeboid morphology (Figure S1A). However, microglia maintained on the astrocyte/neuron co-cultures acquired a more ramified morphology, suggestive of a more mature in-vivo-like phenotype (Figure 1B,C). We next analyzed the influence of neuron-astrocyte co-culture on the microglial transcriptome, as well as well the influence of microglia on the neurons and astrocytes. We performed RNA-seq on (rat) microglia monoculture, (mouse) neuron/(human) astrocyte co-culture, and (mouse) neuron/(human) astrocyte/(rat) microglia triple co-culture. We performed bulk RNA-seq on both co-cultures and monoculture, and all sets of reads were subjected to the same Sargasso workflow (Qiu et al., 2018) to identify unambiguously rat (microglial), mouse (neuronal), and human (astrocytic) reads (Figure S3A). Only ∼3% of reads are lost due to their sequences being 100% conserved between two or more species across the entire paired-end read (Figure S3A). The influence of microglia on astrocytes and neurons was modest, significantly altering the expression of 62 and 8 genes, respectively (Figures S3B and S3C; Table S2), whereas the microglial transcriptome was profoundly altered by the presence of neurons and astrocytes; ∼3,900 genes changes were observed (Figures 1D and S3D; Table S2). Of note, microglial homeostatic signature genes such as Cx3cr1, Tmem119, Tgfbr1, and Sparc, that become downregulated in isolated culture compared to in vivo (Bohlen et al., 2017; Gosselin et al., 2017), are robustly upregulated in the presence of neurons and astrocytes (Figure 1D), validated in the case of Cx3cr1 and Tmem119 by qRT-PCR using species-specific primers (Figure S3E).
We next systematically tested the hypothesis that astrocytes and neurons provide an environment that rescues the loss of microglial homeostatic signature gene (MHSG) expression. We took the set of MHSGs defined by Butovsky et al. (2014) and looked at the influence of neuron-astrocyte co-culture on those MHSGs whose expression is >2-fold lower after maintenance in culture for 7 days relative to immediately post-isolation from the brain, as reported by Gosselin et al. (2017). These genes were induced by neuron-astrocyte co-culture (Figure 1E), evidence that they are sufficient to provide a surrogate CNS microenvironment to maintain MHSG expression. We also tested a panel of MHSGs in a single-species system, mouse microglia in the presence or absence of mouse neuron/astrocyte co-culture (Figure S3F), and validated the upregulation of surface expression of Cx3cr1 protein in the same system by flow cytometry (Figure S3G). Thus, while the mixed-species system enables multiple cell types to be analyzed in parallel (see below), key MHSG expression is also promoted by neuron/astrocyte co-culture in an all-mouse system.
We next sought to define whether the environment provided by astrocyte/neuron co-culture with regard to MHSG induction could be recapitulated by co-culture with astrocytes or neurons alone. Interestingly, the effect of astrocytes alone was weak and that of neurons nonsignificant (Figures 1F and 1G), both weaker than their combined effects (Figures S3H and 1H). This suggests that both neuron- and astrocyte-derived signals are simultaneously required to drive MHSG expression in microglia or that neurons or astrocytes influence the other cell type to provide a single signal.
Certain MHSGs are repressed following inflammatory insults and are part of a wider gene cluster downregulated in models of AD, amyotropic lateral sclerosis (ALS), and multiple sclerosis (MS) as well as age—the so-called MGnD (Krasemann et al., 2017). We observed that neuron/astrocyte co-culture significantly induced expression of the group of genes repressed in the MGnD profile (Figure S4A). As with the MHSG set, neither neurons nor astrocytes alone boosted expression of the gene set repressed in the MGnD profile (Figures S4B and S4C). Thus, neurons and astrocytes boost expression of MHSGs whose expression is repressed ex vivo and also boost expression of an overlapping set of genes whose expression is repressed in age and disease in vivo.
Neurons and astrocytes combine to repress an interferon-related gene cluster
Combined neuron-astrocyte co-cultures induced the downregulation of significant numbers of genes in microglia compared to microglial monocultures (Figure 1D). We therefore investigated whether these changes represent the transformation of microglia to a particular state or the reversal of a particular state ordinarily induced by removal of microglia from their CNS environment. We took advantage of a meta-analysis that identified seven co-regulated gene sets in isolated microglia across a variety of neurodegenerative, inflammatory, neoplastic, and infectious disease models (Friedman et al., 2018). Cross-reference of these gene sets to the data of Gosselin et al. revealed that one, the “interferon-related” gene (IRG) set, became upregulated after maintenance of microglia for 7 days in culture away from their normal CNS microenvironment (data not shown). The IRG set is upregulated in microglia in vivo most strongly in response to viral infection (Friedman et al., 2018). Analysis of our data revealed that members of this IRG set that are ordinarily induced in culture (according to the data of Gosselin et al., 2017) were repressed in microglia by combined neuron-astrocyte co-culture (Figure 2A) and much more modestly influenced by co-culture with neurons or astrocytes alone (Figures 2B, 2C, and S5A). To further define under what circumstances these neuron/astrocyte-repressed IRGs are expressed in vivo, we exploited a single-cell RNA-seq resource that profiled mouse microglia across the lifespan as well as following lysophosphatidylcholine (LPC)-induced white matter injury (Hammond et al., 2019). Mapping these neuron/astrocyte-repressed IRGs (Figure 2A) into this group of 76,000 microglia revealed that they concentrated in a single cluster, cluster 9 (Figures S5B and S5C), noted in the original publication to be “… composed predominantly of microglia from the focal white matter injury” (Hammond et al., 2019). Among the non-injured populations, expression was highest in the microglia of aged (postnatal day 540 [P540]) mice (Figures S5B and S5C). Collectively, these data support a model whereby microglia maintained in isolation away from the CNS microenvironment acquire a type of age- and injury-associated profile, which is repressed by a combination of neuron- and astrocyte-derived signals. A heatmap showing all genes significantly repressed in microglia by co-culture further illustrates the difference between the effect of neurons and astrocytes combined compared to either alone (Figure 2D).
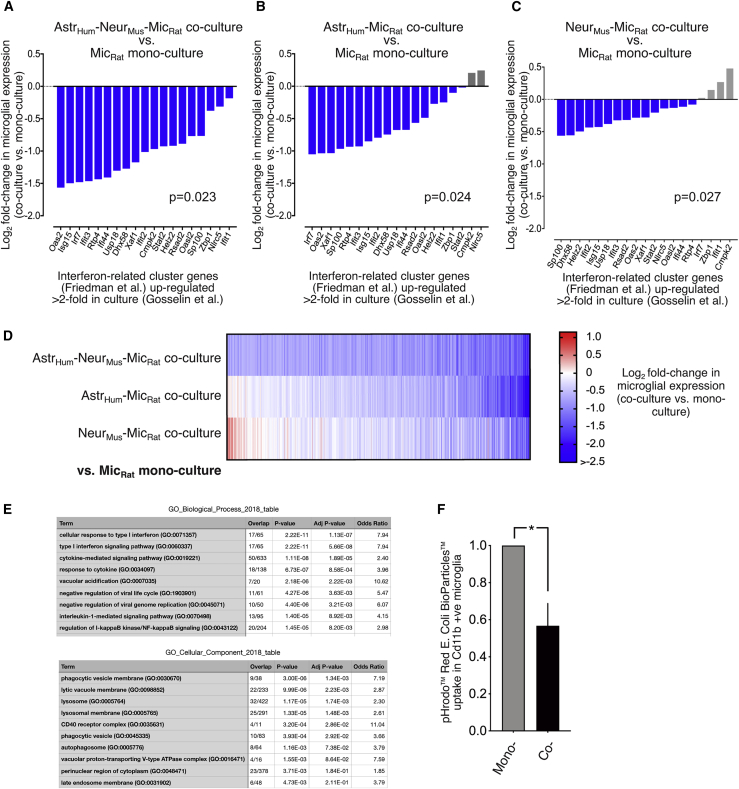
Figure 2.
Neurons and astrocytes combine to repress an IRG cluster
(A–C) Genes considered are those expressed >0.5 FPKM in our data and within the “interferon-related co-regulated genes” defined by Friedman et al. (2018) that are upregulated >2-fold after microglia were maintained in culture for 7 days, compared to their expression immediately post-isolation, according to the data of Gosselin et al. (2017). For each gene, log2FC in microglial gene expression is shown relative to microglial monoculture in microglia cultured with human astrocytes and mouse neurons (A), human astrocyte alone (B), and mouse neurons alone (C). p = 0.023 (A), p = 0.024 (B), p = 0.027 (C), effect of co- versus monoculture conditions, two-way ANOVA.
(D) Heatmap of the log2FC in microglial gene expression in the three different types of co-culture. 602 genes significantly repressed >1.5-fold (DESeq2 P_adj < 0.05) are shown.
(E) The set of genes repressed in microglia by astrocyte/neuron co-culture were subject to ontological analysis and top-ranked GO Biological Processes (top) and Cellular Components (bottom) shown.
(F) A phagocytosis assay was performed on mono- and co-cultured microglia and mean particle uptake calculated. ∗p = 0.007 (n = 5).
Neurons and astrocytes limit microglial phagocytic capacity
In MS patients, microglia near active lesions exhibit a loss of MHSG expression with a concomitant upregulation of phagocytic markers (Zrzavy et al., 2017). In vitro, inflammatory stimulus lipopolysaccharide (LPS) represses MHSG expression and upregulates phagocytic gene expression and phagocytic activity (Lively and Schlichter, 2018; Michaud et al., 2013; Pulido-Salgado et al., 2018). This inverse correlation between MHSG expression and phagocytic activity suggested that neurons and astrocytes may dampen down microglial phagocytic activity, further suggested by ontological analysis of the set of microglial genes repressed by neuron/astrocyte co-culture (Figure 2E). We thus performed a phagocytosis assay on microglia from monocultures and co-cultures using pHrodo Red E.coli BioParticles, which showed that microglial phagocytic capacity was significantly reduced in the presence of neurons and astrocytes (Figure 2F). Thus, in addition to influencing the microglial transcriptome, neurons and astrocytes also influence their functional properties.
Neurons and astrocytes move microglia to a distinct state
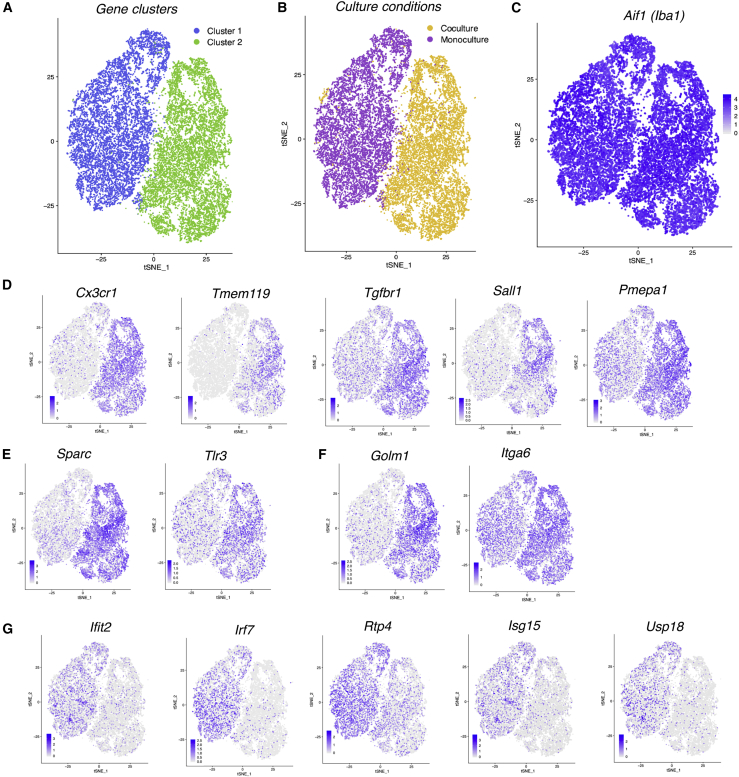
We next investigated the heterogeneity of the response of microglia to neuron/astrocyte co-culture. We fluorescence-activated cell (FAC)-sorted microglia from either monocultures or neuron/astrocyte co-cultures and performed single-cell RNA-seq. An unbiased analysis of the 25,681 microglia sequenced revealed two distinct clusters (Figure 3A), both equally positive for microglial marker Iba1 (Figure 3C). Strikingly, 96.7% of the microglia in cluster 1 were derived from the monoculture and 98.7% in cluster 2 from co-culture (Figure 3B). This indicates that the effect of neuron/astrocyte co-culture is quite uniform, driving nearly all microglia from one state to another. We also mapped onto the clusters a selection of genes common to the MHSG set and the set repressed in the MGnD profile (Figure 3D), as well as genes specific to the MHSG set (Figure 3E) and MGnD-profile-repressed set (Figure 3F), where enrichment in the co-culture-dominated gene cluster 2 can be seen. In contrast, mapping co-culture-repressed IRGs showed enrichment in the monoculture-dominated cluster 1 (Figure 3G), echoing our previous bulk RNA-seq data.
Figure 3.
Single-cell analysis reveals a strong and uniform influence of neurons and astrocytes on the microglial transcriptome
(A–C) Cluster analysis of 25,681 microglial single-cell transcriptomes from monocultures or neuron/astrocyte co-cultures from two independent biological replicates (A) with culture condition (B) and microglial marker Iba1 (C) mapped onto these clusters.
(D–G) Genes mapped into the single-cell data, including a selection of genes common to the MHSG set and the set repressed in the MGnD profile (D), genes specific to the MHSG set (E), and the MGnD-profile-repressed set (F). (G) Selection of IRGs.
TGF-β2 signaling is required for astrocyte/neuron-induction of MHSG expression
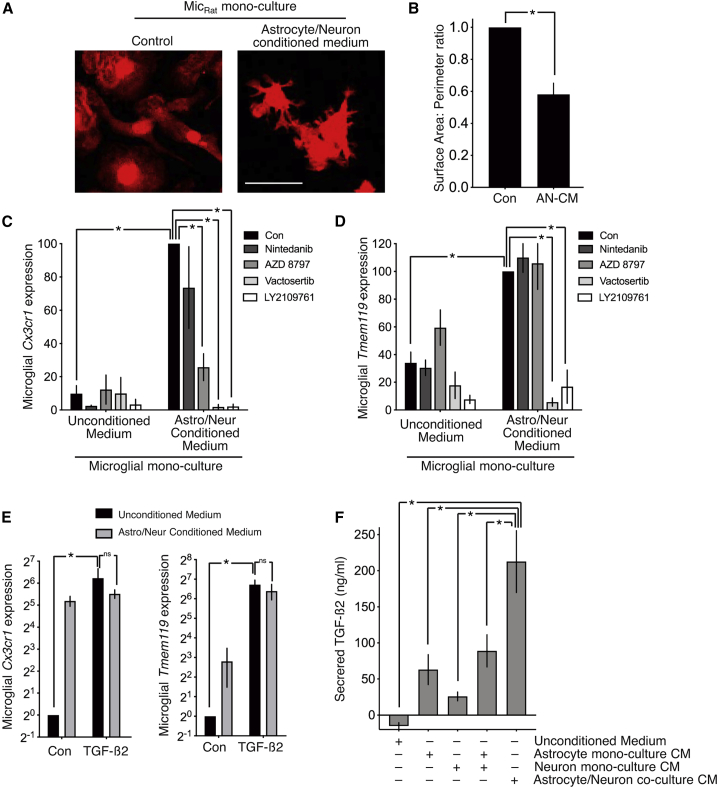
To investigate whether secreted factors released by neurons and/or astrocytes can alter microglial morphology and MHSG expression, microglia monocultures were exposed to medium conditioned by astrocyte/neuron co-culture conditioned medium (AN-CM). Exposure of microglia to AN-CM resulted in an increase in ramification (Figures 4A and 4B) and significant upregulation of a panel of MHSGs tested (Cx3cr1, Tmem119, Cd34, Egr1, Tgfbr1, P2ry13, and Pmepa1; Figure S6A), suggesting that neuron/astrocyte co-cultures are secreting important factors that microglia are unable to make themselves.
Figure 4.
The effects of neurons and astrocytes on microglia are mediated in part by TGF-β2 release
(A) Example images of Iba1-stained monocultures of microglia treated with astrocyte/neuron co-culture conditioned medium (AN-CM) or unconditioned medium. Scale bar, 20 μm.
(B) Surface area/perimeter ratio was calculated in microglial monocultures treated as in (A). ∗p = 0.0094, paired t test (120 cells analyzed per condition across n = 4 biological replicates).
(C and D) Microglial monocultures were exposed ±AN-CM for 72 h, with the indicated drugs added 1 h earlier (nintedanib, 500 nM; AZD8797, 10 μM; vactosertib, 100 μM; LY2109761, 3 μM). RNA was extracted and Cx3cr1 (C) and Tmem119 (D) analyzed by qPCR, normalized to Rpl13a.
(E) Microglial monocultures were treated ±AN-CM ±TGF-β2 (20 ng/mL) and Cx3cr1 and Tmem119 mRNA analyzed as in (C) and (D). ∗p < 0.0001 in all cases, two-way ANOVA plus Dunnett’s post hoc test (n = 3–6). (E) ∗p = 0.010 (Cx3cr1), 0.0006 (Tmem119), two-way ANOVA plus Sidak’s post hoc test (n = 4).
(F) TGF-β2 was measured by ELISA in the indicated media, conditioned for 72 h. ∗p = 0.013, 0.031, 0.028, 0.046, one-way ANOVA plus Sidak’s post hoc (n = 5).
To look for candidates, we performed mass spectrometry analysis on the AN-CM (n = 4) as well as on medium conditioned by monocultured microglia (M-CM) to look for factors present in AN-CM but absent in M-CM. The five highest-expressed candidates (based on label-free quantification intensity [LFQI]) were Cx3cl1 (fractalkine), TGF-β2, Igfbp4, Igfbp5, and Pdgf-A (Table S3). Since Igfbps primarily modulate the actions of Igfs, which were not detected in either M-CM or AN-CM, we focused on the other three candidate ligands. We tested antagonists of the receptors of these ligands for the ability to inhibit the effects of AN-CM in upregulating MHSGs Cx3cr1 and Tmem119 in microglia monocultures. The platelet-derived growth factor receptor/vascular entholelial growth factor receptor (PDGFR/VEGFR) inhibitor nintedanib had no effect on induction of Cx3cr1 or Tmem119, and the Cx3cl1 receptor (Cx3cr1) antagonist AZD8797 reduced induction of Cx3cr1 itself, but not Tmem119 (Figures 4C and 4D). However, inhibition of TGF-βR1/2 signaling with vactosertib (which also inhibits ALK4) or LY2109761 (a specific TGF-βR1/2 antagonist) inhibited the induction of both Cx3cr1 and Tmem119 by AN-CM (Figures 4C and 4D). Moreover, exogenous TGF-β2 was sufficient to induce Cx3cr1 and Tmem119 in microglial monocultures and occlude the effect of AN-CM (Figure 4E). Collectively, these data suggest that TGF-β2 signaling is part of the mechanism by which neurons and astrocytes promote MHSG expression.
We confirmed TGF-β2 presence in AN-CM by ELISA (Figure 4F), comparing to levels in medium conditioned by equal numbers of neurons and astrocytes in separate monocultures (N-CM and A-CM, respectively). The TGF-β2 concentration in AN-CM was higher than that in N-CM and A-CM added together, indicating that neurons and astrocytes cooperate in some way to enhance TGF-β2 production and/or secretion from one or both cell types.
Neurons and astrocytes regulate the microglial response to LPS
Having established that neuron-astrocyte co-culture influences resting microglial profile, we wanted to establish its effect on responses to inflammatory cues. The bacterial endotoxin LPS activates canonical TLR4 signaling in microglia in a dose-dependent manner (Poltorak et al., 1998). We compared the microglial response to two concentrations of LPS when in monoculture with the response when in neuron-astrocyte co-culture. The higher concentration (500 ng/mL, “high LPS”) was chosen to deliver a maximal response and the lower concentration (25 ng/mL, “low LPS”) an intermediate response. We first defined the extent to which LPS-induced changes observed in microglial co-culture resemble those in vivo using two published datasets (Hirbec et al., 2018; Srinivasan et al., 2016). We interrogated genes induced >4-fold by LPS injection in vivo in each study and saw a robust induction of this group of genes by high LPS in microglia co-cultured with neurons and astrocytes (Figures S7A and S7B), confirming that changes observed in the co-culture are relevant to the in vivo response.
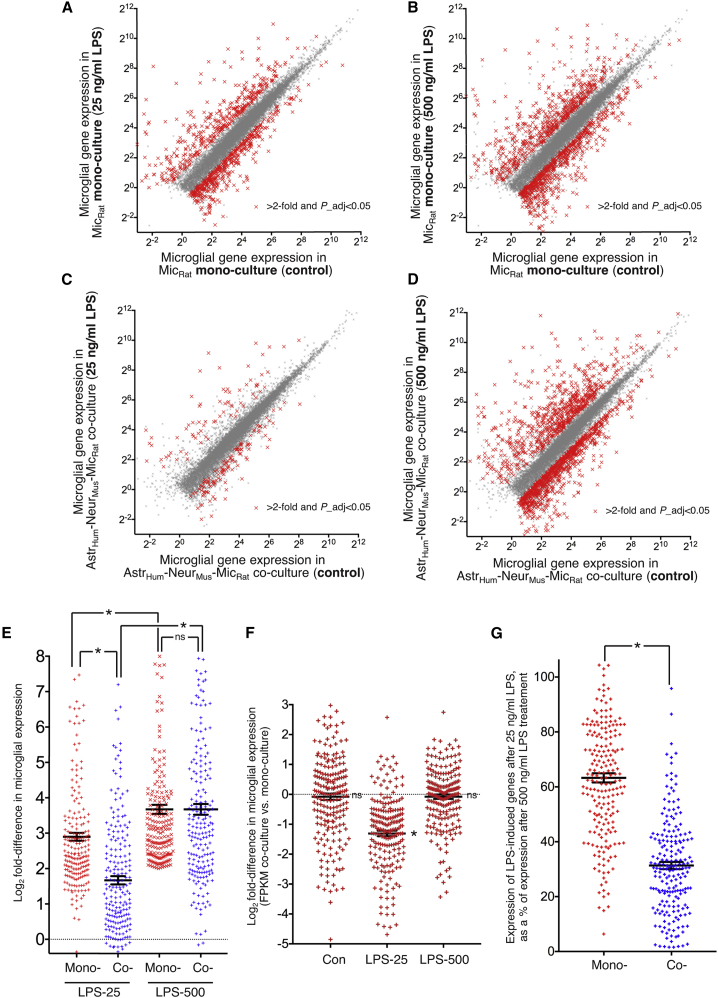
We observed that low LPS induced a robust transcriptional response in microglial monocultures (Figure 5A), with high LPS triggering a slightly stronger response (Figure 5B). Strikingly, however, microglia in neuron-astrocyte co-culture responded weakly to low LPS compared to monoculture (Figure 5C). In contrast, their response to high LPS was strong, not dissimilar to the monocultures response to high LPS (Figure 5D). To assess this quantitatively, we plotted the log2 fold change (log2FC) of genes induced >4-fold (raw fold-change) by high LPS in microglial monocultures, for all conditions (Figure 5E; Table S4). The log2FC of these LPS-response genes by low LPS was lower than high LPS for both co-culture and monoculture (Figure 5E). However, the log2FC by low LPS in monoculture was far higher than the log2FC by low LPS in co-cultured microglia (Figure 5E). Also of note, the log2FC of these LPS-response genes by high LPS showed no difference between monoculture and co-culture (Figure 5E). We also calculated the difference (co-culture versus monoculture) in absolute FPKM levels of LPS-response genes under basal, low LPS and high-LPS conditions (Figure 5F). This revealed an absence of any difference in basal or maximal (high-LPS induced) expression levels, and a clear reduction in expression levels (co-culture versus monoculture) after low-LPS treatment (Figure 5F). Consistent with this, expression levels after low-LPS treatment, compared to high-LPS treatment, were far lower in co-culture, compared to monoculture (Figure 5G). Collectively, this supports a model whereby neurons and astrocytes provide powerful modulatory cues that influence the dose-response of microglia to TLR4 activation, without affecting basal expression of LPS-response genes or the maximal response.
Figure 5.
Neurons and astrocytes regulate the microglial response to LPS
(A–D) LPS-induced microglial gene expression in microglial monoculture (A and B) and co-culture (C and D) in response to 16-h LPS treatment at 25 ng/mL (A and C) or 500 ng/mL (B and D). FPKM of all genes shown that average >1 FPKM across the datasets is plotted for control (x axis) versus LPS (y axis). Highlighted with red crosses are the genes whose expression is significantly changed (DESeq2 P_adj < 0.05) by >2-fold (n = 4).
(E) Log2FC in microglial gene expression is shown in the indicated cultures and in response to the indicated concentrations of LPS. The set of genes analyzed is the 206 induced >4-fold by the high does (500 ng/mL) of LPS in microglial monocultures is shown for all conditions. Mono-, microglial monoculture; Co-, (rat) microglia (mouse) neuron (human) astrocyte co-culture. ∗p values (left to right): <0.0001, 0.0001, <0.0001; ns, >0.9999; two-way ANOVA followed by Tukey’s post hoc test (n = 206).
(F) For the same set of the LPS-responsive genes in (E), we calculated the Log2 fold-change in microglial gene expression in co-culture versus monoculture, under the three different experimental treatments (con, 25 ng/ml LPS, 500 ng/ml LPS). p values (left to right):0.478 (ns), 3.05E-8 (∗), 0.141 (ns); 2-tailed paired Student’s t test on FPKM values in monoculture versus co-culture under each treatment condition.
(G) For the same set of the LPS-responsive genes in (E) and (F), the expression level (FPKM) of each gene under 25-ng/mL LPS conditions was calculated as a percentage of that gene’s expression level under 500-ng/mL LPS conditions. ∗p = 3.01E-39, unpaired two-tailed Student’s t test (n = 206)
Our observations in Figures 1 and 2 suggest that the microglial resting state is modified by a combination of neuron- and astrocyte-mediated effects more than either alone. We therefore asked if this was also the case for their effects on microglial responses to low LPS. Microglia were cultured in the presence or absence of either astrocytes alone or neurons alone and treated with or without low LPS. We plotted the log2FC of genes induced by low LPS in microglial monoculture (x axis) against the corresponding log2FC in microglia in co-culture with astrocytes (Figure S7C), neurons (Figure S7D), and neuron/astrocytes combined (Figure S7E). Linear regression analysis revealed steeper slopes comparing microglial monoculture responses to co-culture with either astrocytes or neurons alone (0.63 and 0.70, respectively; Figures S7C and S7D) than that comparing microglial monoculture responses to co-culture with both neurons and astrocytes (0.4; Figure S7E), consistent with the influence of neurons and astrocytes on microglial LPS responses were stronger when both present.
As noted above, an advantage of the mixed-species co-culture system is that the transcriptomes of all cell types can be analyzed in parallel. While the influence of unchallenged microglia on the transcriptomes of astrocytes and neurons is modest (Figures S3B and S3C), activated microglia release cytokines that act on both neurons and astrocytes. We compared neuron and astrocyte transcriptomes in co-culture (LPS treated) versus astrocyte/neuron/microglia co-culture (LPS treated) to identify the microglia-dependent effects of LPS (distinguishing it from any direct effect of LPS on the neurons or astrocytes). This revealed large numbers of genes both up- and downregulated in astrocytes and neurons due to LPS-activated microglia (Figures 6A and 6B; Table S5). Moreover, a comparison of the microglia-dependent changes induced in astrocytes in the presence of LPS with those induced in astrocytes in vivo after LPS injection (Srinivasan et al., 2016) showed a good agreement between in vitro and in vivo data (Figure S7F). A similar comparison (this time with the changes induced in the neuron/astrocyte/microglia co-culture in the presence or absence of LPS) also showed a good agreement with the in vivo data (Figure S7G). Of note, the astrocytic gene expression changes induced in vivo by LPS injection were not observed in response to LPS treatment of microglia-lacking astrocyte/neuron co-culture (Figure S7H), consistent with the role of microglia in mediating the inflammatory response in astrocytes in vivo.
Figure 6.
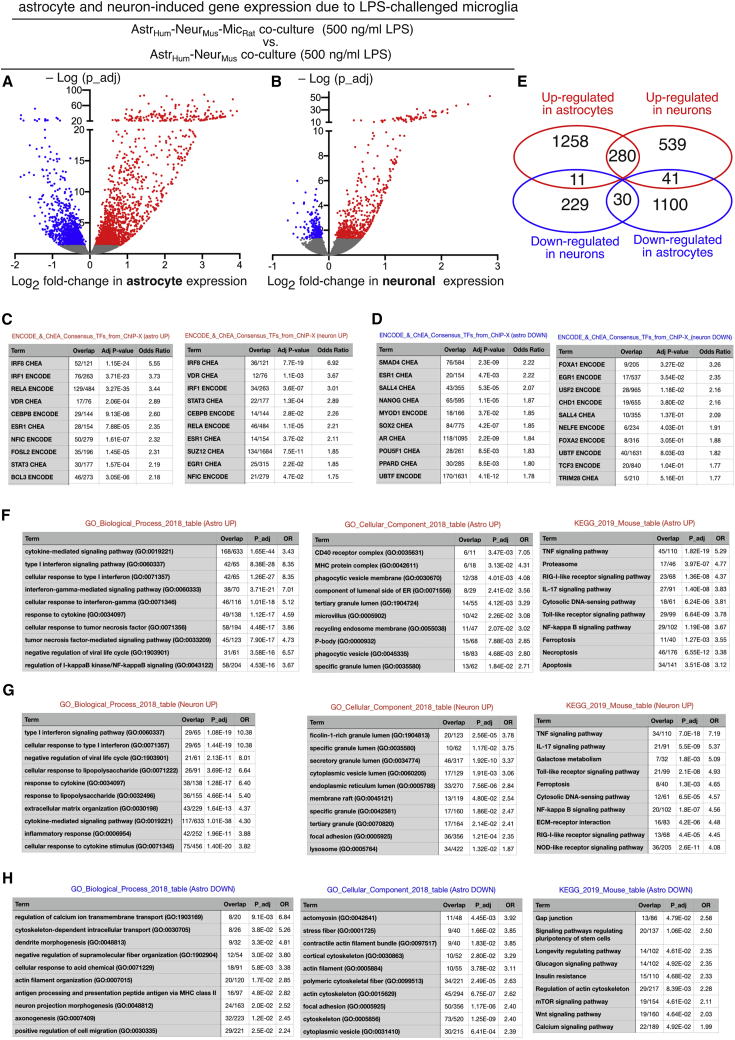
LPS-activated microglia trigger strong responses in astrocytes and neurons
(A and B) Volcano plots of the changes in gene expression in astrocytes (A) and neurons (B) due to microglia when in the presence of LPS (500 ng/mL), identified by applying the Sargasso workflow and sorting the human (astrocyte) reads and mouse (neuron) reads.
(C and D) Genes induced (C) or repressed (D) in astrocytes (left) and neurons (right) by activated microglia were analyzed for enrichment in ENCODE and ChEA Consensus target genes from ChIP-X on the Enrichr platform (Kuleshov et al., 2016). Significantly enriched transcription factor motifs were ranked by fold-enrichment, and the top 10 are shown.
(E) Venn diagram showing the overlap in genes significantly changed in neurons and astrocytes by LPS-activated microglia.
(F–H) Genes induced in astrocytes (F) or neurons (G) and repressed in astrocytes (H) were subject to ontological analysis for enrichment in GO Biological Process (left), GO Cellular Component (middle), and KEGG pathways (right). Enriched terms were ranked by fold enrichment, and the top 10 are shown.
We next performed analyses of the gene sets up- and downregulated in astrocytes and neurons by LPS-activated microglia. Enrichment in Encyclopedia of DNA Elements (ENCODE) and ChIP Enrichment Analysis (ChEA) Consensus target genes from ChIP-X on the Enrichr platform (Kuleshov et al., 2016) revealed putative transcriptional mediators. The genes sets induced in astrocytes and neurons were both enriched in targets of several known mediators of inflammatory responses, including IRF8, IRF1, RELA (NF-κB), STAT3, NFIC, and CEBPβ (Figure 6C). In contrast, there was little similarity in the putative regulators of the downregulated genes (Figure 6D). Consistent with this, there was less overlap in genes repressed by LPS-activated microglia in neurons and astrocytes (only 30) compared to those induced (280; Figure 6E), though the majority remained cell-type specific.
Genes induced in astrocytes and neurons by LPS-activated microglia were enriched in GO Biological Process terms and Kyoto Encyclopedia of Genes and Genomes (KEGG) signaling pathways associated with an inflammatory response (Figures 6F and 6G). GO Cellular Component enrichment analysis revealed a more diverse array of terms, although both gene sets were enriched for components in the phagocytic-endosomal-lysosomal axis (Figures 6F and 6G). Within the set of genes downregulated in astrocytes, changes in cytoskeletal components and pathways were most obvious (Figure 6H), potentially reflecting morphological changes that occur during reactive astrogliosis (Schiweck et al., 2018). Also repressed was the Wnt signaling pathway (including Wnt11, Wnt9a, and Wnt 5b), relevant given its role in neurite outgrowth, synaptogenesis, and synapse maintenance; all functions of astrocytes that are compromised when exposed to LPS-treated microglia (Liddelow and Barres, 2017). Within the group of genes repressed in neurons, little of significance was found, potentially due to a lower number of genes in this set (Figure S7I)
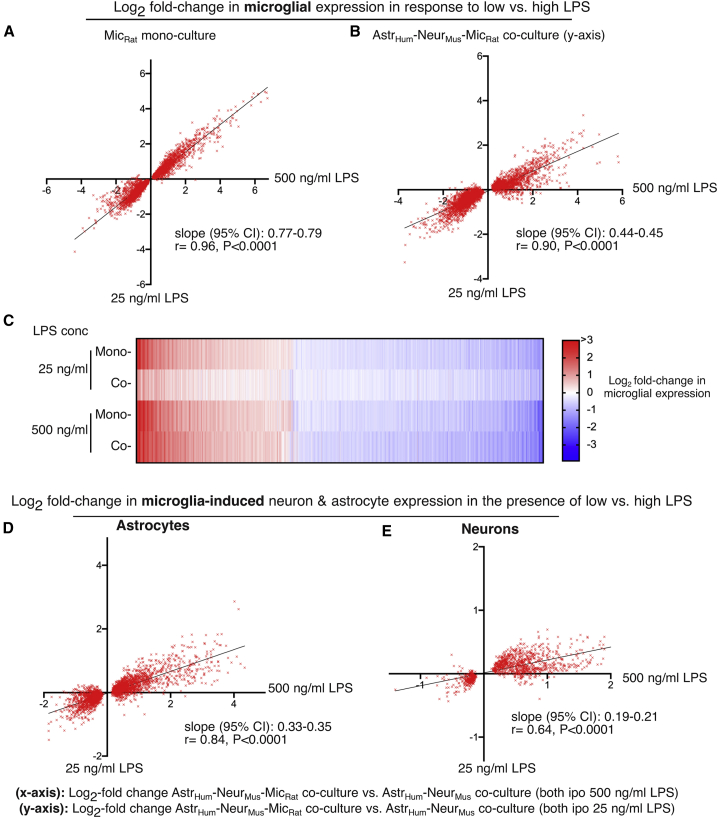
Neurons and astrocytes suppress the response of microglia to a low dose of LPS (25 ng/mL), with little or no impact on the maximal response of microglia (to 500 ng/mL; Figure 5). We wanted to ascertain whether signaling from LPS-challenged microglia to neurons and astrocytes would show a similar LPS dose dependency. We first plotted the log2FC of genes induced in microglia by LPS at a low (y axis) versus high dose (x axis) in monoculture (Figure 7A) and astrocyte/neuron co-culture (Figure 7B). As expected, slope of the graph was far lower (0.45) in co-culture than in monoculture (0.78), since co-culture represses the microglial response at low LPS relative to high LPS, further illustrated by heatmap (Figure 7C). We then performed the same analysis on microglia-dependent changes to astrocytes and neurons in the presence of low versus high LPS. The slopes, 0.34 for astrocytes (Figure 7D) and 0.20 for neurons (Figure 7E), were also low, meaning that in co-culture, microglia challenged with 25 ng/mL LPS induce very modest changes in astrocytes and neurons compared to 500 ng/mL LPS. This is consistent with the effect that astrocytes and neurons themselves have on the microglial response to 25 ng/mL LPS and emphasizes the reciprocal influence that these cell types have on each other.
Figure 7.
The influence of LPS-activated microglia on co-cultured neurons and astrocytes shows a strong LPS dose dependency
(A and B) A comparison of the transcriptional response of microglia to low (25 ng/mL) versus high (500 ng/mL) LPS in monoculture (A) and astrocyte/neuron co-culture (B), focusing solely on significantly changed genes. In addition to the linear regression slope, the correlation coefficient (r) is shown, illustrating the degree of linearity of the relationship between responses to low versus high LPS.
(C) Heatmap of the log2FC in microglial gene expression in monoculture (A) and astrocyte/neuron co-culture at low and high doses of LPS. Genes significantly changed >1.5-fold (DESeq2 P_adj < 0.05) by 500 ng/mL LPS are shown.
(D and E) A comparison of the transcriptional response of astrocytes (D) and neurons (E) to microglia activated by low (25 ng/mL) versus high (500 ng/mL), focusing solely on significantly changed genes. The shallow slopes indicates that the response of astrocytes and neurons to microglia activated by low LPS is smaller than by high LPS, consistent with the dose dependency of the microglia response in co-culture (B).
Discussion
We provide evidence here that neurons and astrocytes combine to specify microglial identity. In doing so, we have described an approach enabling the maintenance of a more mature in-vivo-like microglial profile in culture conditions and the study of immunomodulation due to signaling from other CNS cell types. In conditions devoid of serum at the point of the cells being combined, primary neurons and astrocytes reverse two major shifts in microglial transcriptional profile that ordinarily take place when microglia are kept away from their CNS microenvironment. They signal in a combined manner first to reverse the loss of mature MHSG expression and second to reverse the increase of an infection- and injury-associated IRG set. These shifts are also associated with a return of microglia to their characteristic ramified morphology. The mixed-species approach was central to being able to pick out the changes to microglia and their responses to LPS, within a complex multi-cell-type mixture, with no evidence of innate immune response to the cells due to their distinct species origin.
Surrogate provision of CNS-specific cues
Several studies have reported the rapid (1–2 h) de-differentiation and loss of MHSG expression of microglia in culture (Bohlen et al., 2017; Gosselin et al., 2017). As noted (Bohlen et al., 2017), this means that even acutely isolated microglia poorly resemble the in vivo state, especially as loss of maturity markers will be combined with activated responses associated with the cells’ physical isolation. The loss of microglial signature gene expression can be reversed if microglia are transplanted back into an intact brain (Bohlen et al., 2017), proof that there is no permanent loss of potential of cultured microglia to regain their in vivo profile, given the right cues. Our study shows that neurons and astrocytes combine to provide such cues.
Astrocytes have been shown before to promote a more ramified morphology in cultured microglia (Sievers et al., 1994; Tanaka and Maeda, 1996), and astrocyte-secreted molecules, including TGF-β2, CSF-1, and cholesterol, have been shown to help promote microglial survival in vitro (Bohlen et al., 2017; Butovsky et al., 2014; Gosselin et al., 2017). However, astrocytes are by no means the only CNS cell type involved in microglial regulation; neurons express another CSF1 receptor ligand, interleukin-34 (IL-34), and a plethora of immunomodulatory cell surface proteins, including Cx3cl1, Cd200, Cd47, and Cd22, which bind the microglial receptors Cx3cr1, Cd200r, Cd172, and Cd45, respectively (Hoarau et al., 2011; Perry and Holmes, 2014). Given the variety of secreted and cell surface ligands acting on microglia in vivo, it is perhaps unsurprising that no defined medium is currently able to convincingly promote an in-vivo-like profile in cultured microglia. In this study, we identified TGF-β2 as a secreted factor mediating microglial signature gene induction by astrocyte/neuron co-culture, although it is likely that other factors contribute as well and await discovery. Of note, we did not detect TGF-β1 in AN-CM, but mass spectrometry does not always detect all proteins present.
Our observation that both neurons and astrocytes combined have a more potent influence on microglial gene expression than either cell type alone is consistent with the fact that the aforementioned ligands have different expression profiles in these cells and that cooperation between both cell types may best mimic the CNS microenvironment. This is illustrated by the fact that neuron and astrocyte co-culture augments secreted TGF-β2 production (Figure 4D). Moreover, the combined influence of neurons and astrocytes on microglial LPS responses were also striking, strongly repressing the response of microglia to low concentrations of LPS, without influencing either basal expression of LPS-response genes or maximal LPS-induced levels of these genes. This suggests that rather than exhibiting chronic inflammatory deregulation, isolated microglia are in a primed state, ready to deliver exaggerated responses to relatively mild stimuli. Compared to microglia co-cultured with neurons and astrocytes, microglial monocultures express significantly higher levels of components of inflammatory cascades (including Irak3 and Myd88) and lower levels immunomodulatory receptor Cx3cr1 and Socs6 and Socs2, negative regulators of microglial inflammatory responses (Basrai et al., 2016). Collectively, these and other genes may contribute to this priming effect and suggest that in vivo, neuron- and astrocyte-derived signals may combine to repress this primed state. Of note, microglial priming is thought to be a contributor to pathoprogression in neurodegenerative disease induced by a combination of lowered inhibitory neuronal or glial ligands and misfolded protein aggregate accumulation (Perry and Holmes, 2014). Understanding the mechanisms underlying priming and its suppression may lead to strategies that influence the neuroinflammatory environment, without affecting the maximal range of microglial responses.
A platform for probing inter-cell-type signaling that controls microglial properties
The fact that microglial properties are a function of their surroundings makes this system useful for understanding how other cell-type properties affect microglial function. For example, microglia in vivo have transcriptional profiles that reflect a significant degree of regional heterogeneity, which may be due in part to the differential expression of microglia-influencing ligands in neurons and macroglia in different brain regions. The platform we describe here may be useful in helping to answer questions around the cell-autonomous versus non-cell-autonomous basis for regional microglial heterogeneity. Moreover, given that synaptic activity drives programs of gene expression in neurons themselves and also nearby astrocytes (Bell and Hardingham, 2011; Hasel et al., 2017), the impact of synaptic activity on microglial properties could be explored using this platform. Another advantage of our system is that the genotype of individual cell types can be manipulated or changed independently. For example, mouse knockin and induced pluripotent stem cell (iPSC) studies have shown that the haplotype of the AD risk gene APOE can strongly influence neuroinflammation and microglial properties (Lin et al., 2018; Shi et al., 2017; Tzioras et al., 2018). While some of these may be due to cell-autonomous effects of APOE-expressing microglia, astrocytes are the major APOE-expressing cell in the CNS. One could envisage an experiment whereby the APOE haplotype of iPSC-derived astrocytes is altered and the influence on microglia, both basally and in response to inflammatory insults, is assessed.
More generally, the principles described in this study could be adapted to investigate the regulation of identity of other tissue-resident macrophage populations, which, like microglia, possess unique transcriptional signatures (Gordon et al., 2014; Lavin et al., 2014). These transcriptional signatures are substantially dictated by continued signals from the local tissue microenvironment. Indeed, transplantation studies of peritoneal macrophages into the lung alveolar cavity saw them adopt a profile akin to lung-resident macrophages, indicative of their continued plasticity in terms of their specialism (Lavin et al., 2014). A mixed-species co-culture approach may aid in identifying key determinants of other types of tissue-resident macrophage specialization.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| mouse anti-GFAP | Sigma | Cat# C9205 |
| rabbit anti-Iba1 | Wako Fujifilm | Cat# 019-19741 |
| Cy3-conjugated NeuroChrom | Merck Millipore | Cat#ABN2300C3 |
| mouse anti-Brevican | Sigma | Cat#SAB5200870 |
| Alexa Fluor 647 conjugated anti- CD11b | Biolegend | Cat#101220 |
| PE conjugated anti-mouse CX3CR1 | Biolegend | Cat#149005 |
| Wisteria Floribunda agglutinin | Vector Labs | Cat# B-1355-2 |
| Chemicals, peptides, and recombinant proteins | ||
| Nintedanib | Selleckchem | Cat# S1010 |
| AZD8797 | MedChemExpress | Cat# HY-13848 |
| LY2109761 | Selleckchem | Cat# S2704 |
| TGF-β2 | R&D Systems | Cat# 7346-B2-005 |
| pHrodo™ Red E. Coli BioParticles™ | ThermoFisher | Cat# P35361 |
| Critical Commercial Assays | ||
| High Pure RNA Isolation Kit | Roche | Cat# 11828665001 |
| Transcriptor First Strand cDNA Synthesis Kit | Roche | Cat#04379012001 |
| SYBR Green MasterRox | Roche | Cat# 4913850001 |
| TGF beta-2 Human ELISA Kit | Invitrogen | Cat# BMS254 |
| TruSeq stranded mRNA-seq kit | Illumina | Cat# 20020594 |
| Deposited data | ||
| RNA-seq Data | European Nucleotide Archive | E-MTAB-5987 E-MTAB-7776 E-MTAB-10030 |
| Experimental models: organisms/strains | ||
| Primary Human astrocytes | Caltag Medsystems | Cat#SC-1800 |
| WT CD1 Mice | In-house breeding | N/A |
| Wt Sprague-Dawley Rats | In-house breeding | N/A |
| Oligonucleotides | ||
| qPCR primers | Sigma | See Table in Methods |
| Software and algorithms | ||
| Seurat | PMID:22434839 | SCR_007322 |
| Sargasso | PMID:30250293 | http://statbio.github.io/Sargasso/ |
| MaxQuant | PMID:22434839 | SCR_014485 |
| featureCounts | PMID:22434839 | SCR_012919 |
| STAR | PMID:22434839 | SCR_015899 |
| DESeq2 | PMID:22434839 | SCR_015687 |
| Cell Ranger | PMID:22434839 | SCR_017344 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Giles Hardingham (Giles.Hardingham@ed.ac.uk).
Materials availability
This study did not generate new unique reagents.
Data and code availability
All RNA-seq data that support the findings of this study is available at the European Bioinformatics Institute (ArrayExpress: E-MTAB-5987, ArrayExpress: E-MTAB-7776, and ArrayExpress: E-MTAB-10030). All other data are available from the lead contact upon reasonable request.”
Experimental model and subject details
Generating microglia-neuron-astrocyte co-cultures
For (rat) microglia- (mouse) neuron- (human) astrocyte co-cultures, primary microglia were generated from postnatal rat pups (P0-P2) as described in a recent detailed, open-access protocol (Qiu et al., 2018). Briefly, neocortices were dissected, enzymatically digested, homogenized to single cell suspension and plated down onto poly-D-Lysine-coated flasks. Cultures were maintained for 12 days in DMEM+10% FBS, with medium replacements every 3-4 days. After this period of time, the culture contains predominantly astrocytes and microglia, since the neurons have largely died off under the sub-optimal (for them) culture conditions. To detach the weakly adherent microglia, flasks were placed on an orbital shaker at 250 rpm for 1 h at 37°C, centrifuged to obtain a pellet, then resuspended in Neurobasal A medium (without serum) and counted using a hemocytometer, adjusting the concentration to 2 X 105 cells/ml, ready for seeding onto neuron-astrocyte co-cultures (see below) or onto poly-D-Lysine-coated 24 well plates for generating mono-cultures.
Human astrocytes (Caltag Medsystems cat. no. SC-1800) were maintained on poly-D-lysine-coated tissue culture flasks in Astrocyte Medium (Caltag Medsystems cat. no. SC-1801), and passaged according manufacturer’s instructions. Astrocytes were always used at 7 passages or less and seeded at 3-5 X 105 cells per well of a 24-well plate (poly-D-Lysine-coated). The cells were left until 80%–90% confluent, which typically took 3 days. At this point, primary mouse neocortical neurons were prepared as described (Qiu et al., 2018) from E17.5 CD1 mice and seeded at 1 X 105 cells per well onto the astrocyte monolayer. These co-cultures were maintained for 7 days in Neurobasal A medium, plus 1% rat serum, in the presence of AraC to prevent any mouse astrocyte proliferation, and any further human astrocyte proliferation. At this point, microglia were seeded onto the co-culture at 2 X 105 cells per well and maintained for 3 further days in Neurobasal A medium. 3 days after microglial seeding, microglia mono-cultures or microglia-neuron-astrocyte co-cultures were transferred to basal TMo medium (Qiu et al., 2018) overnight, prior to treatment with LPS for 16 hours. At this point, RNA was extracted and RNA integrity checked using an Agilent 2100 Bioanalyzer.
For (mouse) microglia-(rat) neuron- (human) astrocyte co-culture, the same workflow as (rat) microglia-(mouse) neuron- (human) astrocyte co-culture was applied, apart from primary microglia were generated from postnatal mouse pups (P0-P2) and primary neurons from E20.5 Sprague Dawley rat embryos. For single-species mouse or rat microglia-neuron-astrocyte co-cultures, the same workflow as above was also applied, except that the astrocytes were cultured from either E17.5 CD1 mouse or E20.5 Sprague Dawley rat embryos respectively, as previously described (Baxter et al., 2011).
Method details
Phagocytosis assay
The (rat) microglial mono-cultures and (rat) microglia- (mouse) neuron- (human) astrocyte co-cultures were generated in the same manner as described above. 3 days after microglial seeding, the media was replaced with pHrodo™ Red E. Coli BioParticles™ conjugate suspension (1:10 dilution in PBS) prepared following manufacturer’s instructions (ThermoFisher) and then incubated in a CO2-free incubator at 37°C for 1 h. The cells were then physically dissociated from the culture dishes, stained with Alexa Flour conjugated 647 anti-rat CD11b antibody (Biolegend) and analyzed on a BD Accuri™ C6 flow cytometer (BD biosciences).
ELISA
Mouse neurons, astrocytes, or co-cultures were prepared as described with fresh Neurobasal A media applied at DIV 4 and harvested 3 days later. TGF-β2 levels in culture medium were assayed using the TGF beta-2 Human ELISA Kit (Invitrogen) as per manufacturer’s instructions.
Cx3cr1 flow cytometry assay
The (mouse) microglial mono-cultures and (mouse) microglia- (mouse) neuron- (mouse) astrocyte co-cultures was generated in the same manner as described above. 3 days after microglial seeding, the cells were then physically dissociated from the culture dishes and stained with Alexa Fluor 647 conjugated anti-mouse CD11b (Biolegend) and PE conjugated anti-mouse CX3CR1 (Biolegend) before analysis on a BD Accuri™ C6 flow cytometer (BD biosciences).
Conditioning the medium
To prepare conditioned medium (human) astrocyte and (mouse) neuron co-cultures were generated as previously described, except that at DiV 4, cells were washed once and media replaced with Neurobasal-A medium in the absence of serum. At DiV 7, media was harvested and co-cultures discarded. Flasks of (rat) microglia, prepared as described above, were detached, centrifuged and pellets were resuspended in either naive Neurobasal-A medium, or conditioned medium, with cells diluted to 2 X 105 cells/ml, and plated in 24 well plates. The following drug treatments were added at plate-down: Nintedanib (500 nM, Selleckchem), AZD8797 (10 μM, Selleckchem), Vactosertib (100 μM, Selleckchem), LY2109761 (3 μM, Selleckchem) mouse TGF-β2 (20 ng/ml, R&D Systems). Microglia were incubated for 72 h, after which they were processed for either immunohistochemistry or RNA isolation and qPCR.
Immunocytochemistry and morphometric quantification
For cell culture immunohistochemistry, established protocols were employed (Baxter et al., 2011; Puddifoot et al., 2012). Briefly, cells were fixed in 4% formaldehyde for 20 min at room temperature, washed with PBS and permeabilized with the detergent NP40 (Life Technologies). Cells were subsequently incubated in primary antibody over night at 4°C. The next day, cells were washed with PBS and incubated with the appropriate Alexa Fluor-conjugated secondary antibody (1:250, ThermoFisher) at room temperature for 1h. Cells were then mounted using the mounting medium Vectashield (Vector Labs). Primary antibodies used include: mouse anti-GFAP (1:400, Sigma), mouse anti-Iba1 (1:750, Abcam), rabbit anti-Iba1 (1:1000, Wako Fujifilm), Cy3-conjugated NeuroChrom (1:500, Merck Millipore), mouse anti-Brevican (1:500, Sigma), biotinylated Wisteria Floribunda agglutinin (1:400, Vector Labs). Images were prepared on a Leica AF6000 using a DFC30X digital camera. For morphometric quantification of microglia, cultures were fixed as above and probed for Iba1. Images were taken using a Nikon A1R confocal microscope. Cell outlines were traced and perimeter/area calculated using ImageJ, with 120 cells measured per condition.
Mass spectrometry
The (rat) microglial mono-cultures and (rat) microglia- (rat) neuron- (rat) astrocyte co-cultures were generated with the same workflow as described above, apart from that microglial mono-cultures and microglia-neuron-astrocyte cocultures were grown in T75 flasks. 3 days after microglial seeding, the flasks were washed with phenol red free TMO and medium was collected 24 h later (n = 4 independent biological replicates). A proteinase inhibitor cocktail (Roche, 11836153001) was dissolved directly into the media before centrifugation at 3000 g for 20 minutes, following which the supernatant was passed through a 0.22 μm filter and then concentrated with Amicon Ultra filters with a 10kDa NMWL (Merk, UFC901024, UFC501096) according to the manufacturer instructions. The samples were denatured with 6M guanidine hydrochloride and then treated with 5mM tris (2-carboxyethyl) phosphine (TCEP) and 10mM chloroacetaldehyde (CAA) for reduction and alkylation respectively, before heating at 95°C for 5 minutes. After cooling to room temperature, the samples were digested first with 0.2 μg endoproteinase LysC in 3M guanidine at 37°C overnight and then with 0.3 μg trypsin in 1M guanidine at 37°C for 4 hours. The resulting peptides were cleaned up using C18 Stage Tips (Rappsilber et al., 2003), separated with an Ultimate 3000-series RSLC Nano System (Thermo Fisher), ionised with an IonOpticks Aurora C18 nano packed emitter in a Proxeon nano source (Thermo Fisher), analyzed on a Orbitrap Fusion Lumos Tribrid Mass Spectrometer (Thermo Fisher), and identified and counted using MaxQuant platform (version 1.6.7.0) and Uniprot proteomics database (Cox and Mann, 2008; UniProt Consortium, 2019).
RNA-seq and species-specific sorting of mixed species reads
To generate RNA-seq data, barcoded RNA-seq libraries were prepared by Edinburgh Genomics using the Illumina TruSeq stranded mRNA-seq kit, according to the manufacturer’s protocol (Illumina). The libraries were pooled and sequenced to 75 base paired-end on an Illumina NovaSeqTM 6000. For single-species RNA-seq experiments sequencing was performed to a depth of approximately 50 million paired-end reads per sample, whereas for mixed species RNA-seq a greater depth of approximately 100 million (for two species), and 150 million (for three species) paired-end reads per sample was done. Species-specific separation of RNA-seq reads was performed using version 1.2 of Sargasso (Qiu et al., 2018) (during which reads were mapped to the mouse, rat and human genomes using version 2.5.3a of STAR (Dobin et al., 2013)). The protocol is described in detail elsewhere (Qiu et al., 2018). Subsequently, per-gene read counts were summarized using featureCounts version 1.5.2. For read mapping and feature counting, genome sequences and gene annotations were downloaded from Ensembl version 94. Differential expression (DGE) analysis on datasets was performed using DESeq2 (Love et al., 2014) (R package version 1.18.1) using a significance threshold set at a Benjamini-Hochberg-adjusted p value of 0.05. Before arriving at a final DGE dataset for the microglia, we carried out an additional control (as recommended in our published protocol (Qiu et al., 2018)) by performing RNA-seq on a two-species co-culture of mouse neurons and human astrocytes, and determining whether the Sargasso workflow resulted in any human or mouse reads being incorrectly called as rat. We took a conservative approach and discarded any genes for which we estimated > 10% of rat reads within the mixed species co-culture could be due to incorrectly called human or mouse reads.
Single-cell RNA-seq
The (rat) microglial mono-cultures and (rat) microglia- (mouse) neuron- (human) astrocyte co-cultures were generated in the same manner as described above. 3 days after microglial seeding to T-25 flasks, the cells were then physically dissociated from the culture dishes in ice-cold dPBS with sodium pyruvate (ThermoFisher) + 0.4% BSA and stained with PE conjugated anti-mouse CD11b (Biolegend). A pure single-cell suspension of microglia was sorted using a BD Aria FACS system. Suspension sufficient to generate 5000 single cell GEMs (gel beads in emulsion) per sample was loaded into a Chromium controller (10X Genomics) in a Single Cell 3′ Chip. Libraries were prepared using the Chromium Single Cell 3′ GEM, Library and Gel Bead Kit v3 (10X Genomics) as per manufacturer’s instructions, with libraries quantified using a DNA 1000 chip (Agilent Technologies). Samples were loaded onto the Illumina NextSeq, with an average of 100 million 75 base-pair reads per sample obtained. Cell Ranger (version 3.1.0), a set of 10X Genomics pipelines to process Chromium single cell data, was used to demultiplex raw base call files to generate forward, reverse and index FASTQ files, and align reads against the mouse reference genome (mm10) to produce count matrices. Following alignment, count matrices per library were aggregated to generate a single counts matrix for downstream analysis. Further analysis was performed using Seurat version 3.1.1 (Stuart et al., 2019). The aggregated counts matrix produced by Cell Ranger was imported into Seurat and pre-processed, removing cells with fewer than 1000 features, and retaining only features which were detected in at least 5 cells. Count data was normalized by Log1p transforming counts which have been multiplied by a scaling factor and scaled to the cell count total. Variance stabilizing transformation was used to identify the top 2000 most variable features. The effect of number of UMIs and percent mitochondrial RNA per cell was regressed out from the counts of the most variable features computed in the previous step, before scaling and centering the data. Clusters were identified by constructing a shared nearest neighbor graph (SNN) from the top 10 principal components computed from the scaled data. In order to test whether microglia in monoculture and coculture clustered along treatment lines and showed a consistently divergent expression profile, the resolution value used when identifying clusters within the SNN was lowered until cells formed two clusters, using a resolution value of 0.02. TSNE dimensionality reduction was used to compare the profile and distribution of the two clusters to that of mono-culture and co-culture libraries respectively to confirm whether they occupied similar cluster space.
RNA extraction, RT-PCR and qPCR
Total RNA extraction from cultures was performed using the High Pure RNA Isolation Kit (Roche) and cDNA was subsequently created using the Transcriptor First Strand cDNA Synthesis Kit (Roche) using the following program: 10 min at 25 °C, 30 min at 55 °C and 5 min at 85 °C. qPCRs were run on a Stratagene Mx3000P QPCR System (Agilent Technologies) using SYBR Green MasterRox (Roche) with 6 ng of cDNA per well of a 96-well plate, using the following program: 10 min at 95 °C, 40 cycles of 30 s at 95 °C, 40 s at 60 °C and 30 s at 72 °C, with a subsequent cycle of 1 min at 95 °C and 30 s at 55 °C ramping up to 95 °C over 30 s (to measure the dissociation curve). Primers were designed to only amplify targets from a single species, and were validated by qPCRs with cDNA generated from pure mouse, rat or human cultures. The following primers were used:
| Species | Gene | Forward primer | Reverse primer |
|---|---|---|---|
| Mouse | Cx3cr1 | CCTGGTGGTCTTTGCC | GCACTTCCTATACAGGTGTCC |
| Tmem119 | CCCACACCGGAGAGAC | CAGGGAACGAGGATGG | |
| Cd34 | CTTCCCCAACTGGCATAC | ATACCCTGGGCCAACC | |
| Tgfbr1 | CATAGTGATACCAATCCCCAG | TGGTGTTTCTGACGTATGAAAC | |
| Pmepa1 | ATGGTGATGGTGGTTATGATTAC | CCTGACACCGTACTCTCTGAG | |
| P2yr13 | CGTCTACCTCAAGAACACTCTG | ATGATGATCTTGAGGAACCTG | |
| Egr1 | CTGCCTCTTCACTCTCTTCTTAC | TGGAACGGAGGCAAAG | |
| Rpl13a | GATGAATACCAACCCCTCC | CGAACAACCTTGAGAGCAG | |
| Rat | Cx3cr1 | AACGAATGTTTGGGTGATTAC | AAGAAGACAACAACCACCAAG |
| Tmem119 | TTCAGTCCCACACCAGG | GAGGACGGGTAGTAGGCTG | |
| Cd34 | ATTCACCTAATAATGTCAGCTCTG | ACACTAGTACCAGCGTCGG | |
| Tgfbr1 | TGAGCGCTGTTAACATCTTATC | GATGACAGCACAAGAGCG | |
| Pmepa1 | CTGGAGCTGAACCGAGAG | CTCACTGTAGGTGGGAGGAG | |
| P2yr13 | CAAGATCGTCGTACCGTTTAG | ACTACTTGATGCCACAGCAG | |
| Egr1 | GTCGTGGCCTCCTCAG | CTATGCCTCCGTCCCA | |
| Rpl13a | CGCACAAGACCAAAAGAG | GTTTCCTTAGCCTCAAGAGC |
Quantification and statistical analysis
Statistical testing of the RNA-seq data is described in that section. Other testing was performed in Prism or Excel and involved a 2-tailed paired Student’s t test, or a one- or two-way ANOVA followed by Sidak’s post hoc test, as indicated in the legends. For t tests, variance was generally found to be similar, abrogating the need for Welsh’s Correction. Error bars indicate the SEM throughout. Throughout the manuscript, independent biological replicates are defined as independently performed experiments on material derived from different animals.
Acknowledgments
This work is funded by UK Dementia Research Institute partner funders the Medical Research Council, Alzheimer’s Research UK, and Alzheimer's Society, as well as the Wellcome Trust and the Simons Initiative for the Developing Brain. We acknowledge the expertise and assistance of Edinburgh Genomics (RNA-seq) and the IGMM Mass Spectrometry facility.
Author contributions
J.Q. and P.S.B. developed and validated the three-species co-culture systems. P.S.B., J.Q., and S.M. performed the experiments. O.D., P.S.B., S.M., X.H., J.Q., K.E., and G.E.H. analyzed the data. J.Q. and G.E.H. conceived and directed the project. G.E.H. and J.Q. wrote the manuscript. Other authors provided critical feedback on the manuscript.
Declaration of interests
The authors declare no competing interests.
Published: March 23, 2021
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2021.108882.
Contributor Information
Giles E. Hardingham, Email: giles.hardingham@ed.ac.uk.
Jing Qiu, Email: jing.qiu@ed.c.uk.
Supplemental information
References
- Basrai H.S., Christie K.J., Turbic A., Bye N., Turnley A.M. Suppressor of Cytokine Signaling-2 (SOCS2) Regulates the Microglial Response and Improves Functional Outcome after Traumatic Brain Injury in Mice. PLoS ONE. 2016;11:e0153418. doi: 10.1371/journal.pone.0153418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baxter P.S., Martel M.A., McMahon A., Kind P.C., Hardingham G.E. Pituitary adenylate cyclase-activating peptide induces long-lasting neuroprotection through the induction of activity-dependent signaling via the cyclic AMP response element-binding protein-regulated transcription co-activator 1. J. Neurochem. 2011;118:365–378. doi: 10.1111/j.1471-4159.2011.07330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell K.F., Hardingham G.E. The influence of synaptic activity on neuronal health. Curr. Opin. Neurobiol. 2011;21:299–305. doi: 10.1016/j.conb.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohlen C.J., Bennett F.C., Tucker A.F., Collins H.Y., Mulinyawe S.B., Barres B.A. Diverse Requirements for Microglial Survival, Specification, and Function Revealed by Defined-Medium Cultures. Neuron. 2017;94:759–773.e8. doi: 10.1016/j.neuron.2017.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butovsky O., Jedrychowski M.P., Moore C.S., Cialic R., Lanser A.J., Gabriely G., Koeglsperger T., Dake B., Wu P.M., Doykan C.E. Identification of a unique TGF-β-dependent molecular and functional signature in microglia. Nat. Neurosci. 2014;17:131–143. doi: 10.1038/nn.3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman B.A., Srinivasan K., Ayalon G., Meilandt W.J., Lin H., Huntley M.A., Cao Y., Lee S.H., Haddick P.C.G., Ngu H. Diverse Brain Myeloid Expression Profiles Reveal Distinct Microglial Activation States and Aspects of Alzheimer’s Disease Not Evident in Mouse Models. Cell Rep. 2018;22:832–847. doi: 10.1016/j.celrep.2017.12.066. [DOI] [PubMed] [Google Scholar]
- Gordon S., Plüddemann A., Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol. Rev. 2014;262:36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosselin D., Skola D., Coufal N.G., Holtman I.R., Schlachetzki J.C.M., Sajti E., Jaeger B.N., O’Connor C., Fitzpatrick C., Pasillas M.P. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356:eaal3222. doi: 10.1126/science.aal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J. Single-Cell RNA Sequencing of Microglia throughout the Mouse Lifespan and in the Injured Brain Reveals Complex Cell-State Changes. Immunity. 2019;50:253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasel P., Dando O., Jiwaji Z., Baxter P., Todd A.C., Heron S., Márkus N.M., McQueen J., Hampton D.W., Torvell M. Neurons and neuronal activity control gene expression in astrocytes to regulate their development and metabolism. Nat. Commun. 2017;8:15132. doi: 10.1038/ncomms15132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirbec H., Marmai C., Duroux-Richard I., Roubert C., Esclangon A., Croze S., Lachuer J., Peyroutou R., Rassendren F. The microglial reaction signature revealed by RNAseq from individual mice. Glia. 2018;66:971–986. doi: 10.1002/glia.23295. [DOI] [PubMed] [Google Scholar]
- Hoarau J.J., Krejbich-Trotot P., Jaffar-Bandjee M.C., Das T., Thon-Hon G.V., Kumar S., Neal J.W., Gasque P. Activation and control of CNS innate immune responses in health and diseases: a balancing act finely tuned by neuroimmune regulators (NIReg) CNS Neurol. Disord. Drug Targets. 2011;10:25–43. doi: 10.2174/187152711794488601. [DOI] [PubMed] [Google Scholar]
- Krasemann S., Madore C., Cialic R., Baufeld C., Calcagno N., El Fatimy R., Beckers L., O’Loughlin E., Xu Y., Fanek Z. The TREM2-APOE Pathway Drives the Transcriptional Phenotype of Dysfunctional Microglia in Neurodegenerative Diseases. Immunity. 2017;47:566–581.e9. doi: 10.1016/j.immuni.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44(W1):W90. doi: 10.1093/nar/gkw377. 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavin Y., Winter D., Blecher-Gonen R., David E., Keren-Shaul H., Merad M., Jung S., Amit I. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liddelow S.A., Barres B.A. Reactive Astrocytes: Production, Function, and Therapeutic Potential. Immunity. 2017;46:957–967. doi: 10.1016/j.immuni.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Lin Y.T., Seo J., Gao F., Feldman H.M., Wen H.L., Penney J., Cam H.P., Gjoneska E., Raja W.K., Cheng J. APOE4 Causes Widespread Molecular and Cellular Alterations Associated with Alzheimer’s Disease Phenotypes in Human iPSC-Derived Brain Cell Types. Neuron. 2018;98:1141–1154.e7. doi: 10.1016/j.neuron.2018.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lively S., Schlichter L.C. Microglia Responses to Pro-inflammatory Stimuli (LPS, IFNγ+TNFα) and Reprogramming by Resolving Cytokines (IL-4, IL-10) Front. Cell. Neurosci. 2018;12:215. doi: 10.3389/fncel.2018.00215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel M.A., Ryan T.J., Bell K.F., Fowler J.H., McMahon A., Al-Mubarak B., Komiyama N.H., Horsburgh K., Kind P.C., Grant S.G. The subtype of GluN2 C-terminal domain determines the response to excitotoxic insults. Neuron. 2012;74:543–556. doi: 10.1016/j.neuron.2012.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matcovitch-Natan O., Winter D.R., Giladi A., Vargas Aguilar S., Spinrad A., Sarrazin S., Ben-Yehuda H., David E., Zelada González F., Perrin P. Microglia development follows a stepwise program to regulate brain homeostasis. Science. 2016;353:aad8670. doi: 10.1126/science.aad8670. [DOI] [PubMed] [Google Scholar]
- Michaud J.P., Hallé M., Lampron A., Thériault P., Préfontaine P., Filali M., Tribout-Jover P., Lanteigne A.M., Jodoin R., Cluff C. Toll-like receptor 4 stimulation with the detoxified ligand monophosphoryl lipid A improves Alzheimer’s disease-related pathology. Proc. Natl. Acad. Sci. USA. 2013;110:1941–1946. doi: 10.1073/pnas.1215165110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimmerjahn A., Kirchhoff F., Helmchen F. Resting microglial cells are highly dynamic surveillants of brain parenchyma in vivo. Science. 2005;308:1314–1318. doi: 10.1126/science.1110647. [DOI] [PubMed] [Google Scholar]
- Okaty B.W., Sugino K., Nelson S.B. Cell type-specific transcriptomics in the brain. J. Neurosci. 2011;31:6939–6943. doi: 10.1523/JNEUROSCI.0626-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okaty B.W., Sugino K., Nelson S.B. A quantitative comparison of cell-type-specific microarray gene expression profiling methods in the mouse brain. PLoS ONE. 2011;6:e16493. doi: 10.1371/journal.pone.0016493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadia S., Soriano F.X., Léveillé F., Martel M.A., Dakin K.A., Hansen H.H., Kaindl A., Sifringer M., Fowler J., Stefovska V. Synaptic NMDA receptor activity boosts intrinsic antioxidant defenses. Nat. Neurosci. 2008;11:476–487. doi: 10.1038/nn2071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perry V.H., Holmes C. Microglial priming in neurodegenerative disease. Nat. Rev. Neurol. 2014;10:217–224. doi: 10.1038/nrneurol.2014.38. [DOI] [PubMed] [Google Scholar]
- Poltorak A., He X., Smirnova I., Liu M.Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Puddifoot C., Martel M.A., Soriano F.X., Camacho A., Vidal-Puig A., Wyllie D.J., Hardingham G.E. PGC-1α negatively regulates extrasynaptic NMDAR activity and excitotoxicity. J. Neurosci. 2012;32:6995–7000. doi: 10.1523/JNEUROSCI.6407-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Salgado M., Vidal-Taboada J.M., Barriga G.G., Solà C., Saura J. RNA-Seq transcriptomic profiling of primary murine microglia treated with LPS or LPS + IFNγ. Sci. Rep. 2018;8:16096. doi: 10.1038/s41598-018-34412-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu J., Dando O., Baxter P.S., Hasel P., Heron S., Simpson T.I., Hardingham G.E. Mixed-species RNA-seq for elucidation of non-cell-autonomous control of gene transcription. Nat. Protoc. 2018;13:2176–2199. doi: 10.1038/s41596-018-0029-2. [DOI] [PubMed] [Google Scholar]
- Ransohoff R.M. How neuroinflammation contributes to neurodegeneration. Science. 2016;353:777–783. doi: 10.1126/science.aag2590. [DOI] [PubMed] [Google Scholar]
- Rappsilber J., Ishihama Y., Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal. Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- Salter M.W., Stevens B. Microglia emerge as central players in brain disease. Nat. Med. 2017;23:1018–1027. doi: 10.1038/nm.4397. [DOI] [PubMed] [Google Scholar]
- Schiweck J., Eickholt B.J., Murk K. Important Shapeshifter: Mechanisms Allowing Astrocytes to Respond to the Changing Nervous System During Development, Injury and Disease. Front. Cell. Neurosci. 2018;12:261. doi: 10.3389/fncel.2018.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y., Yamada K., Liddelow S.A., Smith S.T., Zhao L., Luo W., Tsai R.M., Spina S., Grinberg L.T., Rojas J.C., Alzheimer’s Disease Neuroimaging Initiative ApoE4 markedly exacerbates tau-mediated neurodegeneration in a mouse model of tauopathy. Nature. 2017;549:523–527. doi: 10.1038/nature24016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sievers J., Parwaresch R., Wottge H.U. Blood monocytes and spleen macrophages differentiate into microglia-like cells on monolayers of astrocytes: morphology. Glia. 1994;12:245–258. doi: 10.1002/glia.440120402. [DOI] [PubMed] [Google Scholar]
- Slaker M.L., Harkness J.H., Sorg B.A. A standardized and automated method of perineuronal net analysis using Wisteria floribunda agglutinin staining intensity. IBRO Rep. 2016;1:54–60. doi: 10.1016/j.ibror.2016.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K., Friedman B.A., Larson J.L., Lauffer B.E., Goldstein L.D., Appling L.L., Borneo J., Poon C., Ho T., Cai F. Untangling the brain’s neuroinflammatory and neurodegenerative transcriptional responses. Nat. Commun. 2016;7:11295. doi: 10.1038/ncomms11295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stansley B., Post J., Hensley K. A comparative review of cell culture systems for the study of microglial biology in Alzheimer’s disease. J. Neuroinflammation. 2012;9:115. doi: 10.1186/1742-2094-9-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive Integration of Single-Cell Data. Cell. 2019;177:1888–1902 e1821. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka J., Maeda N. Microglial ramification requires nondiffusible factors derived from astrocytes. Exp. Neurol. 1996;137:367–375. doi: 10.1006/exnr.1996.0038. [DOI] [PubMed] [Google Scholar]
- Tzioras M., Davies C., Newman A., Jackson R., Spires-Jones T. Invited Review: APOE at the interface of inflammation, neurodegeneration and pathological protein spread in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2018;45:327–346. doi: 10.1111/nan.12529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- UniProt Consortium UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res. 2019;47(D1):D506–D515. doi: 10.1093/nar/gky1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Brink S.C., Sage F., Vértesy Á., Spanjaard B., Peterson-Maduro J., Baron C.S., Robin C., van Oudenaarden A. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods. 2017;14:935–936. doi: 10.1038/nmeth.4437. [DOI] [PubMed] [Google Scholar]
- Wu Y., Dissing-Olesen L., MacVicar B.A., Stevens B. Microglia: Dynamic Mediators of Synapse Development and Plasticity. Trends Immunol. 2015;36:605–613. doi: 10.1016/j.it.2015.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wylot B., Konarzewska K., Bugajski L., Piwocka K., Zawadzka M. Isolation of vascular endothelial cells from intact and injured murine brain cortex-technical issues and pitfalls in FACS analysis of the nervous tissue. Cytometry A. 2015;87:908–920. doi: 10.1002/cyto.a.22677. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Sloan S.A., Clarke L.E., Caneda C., Plaza C.A., Blumenthal P.D., Vogel H., Steinberg G.K., Edwards M.S., Li G. Purification and Characterization of Progenitor and Mature Human Astrocytes Reveals Transcriptional and Functional Differences with Mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zrzavy T., Hametner S., Wimmer I., Butovsky O., Weiner H.L., Lassmann H. Loss of ‘homeostatic’ microglia and patterns of their activation in active multiple sclerosis. Brain. 2017;140:1900–1913. doi: 10.1093/brain/awx113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All RNA-seq data that support the findings of this study is available at the European Bioinformatics Institute (ArrayExpress: E-MTAB-5987, ArrayExpress: E-MTAB-7776, and ArrayExpress: E-MTAB-10030). All other data are available from the lead contact upon reasonable request.”