Figure 1.
Neurons and astrocytes combine to drive microglial homeostatic signature gene expression
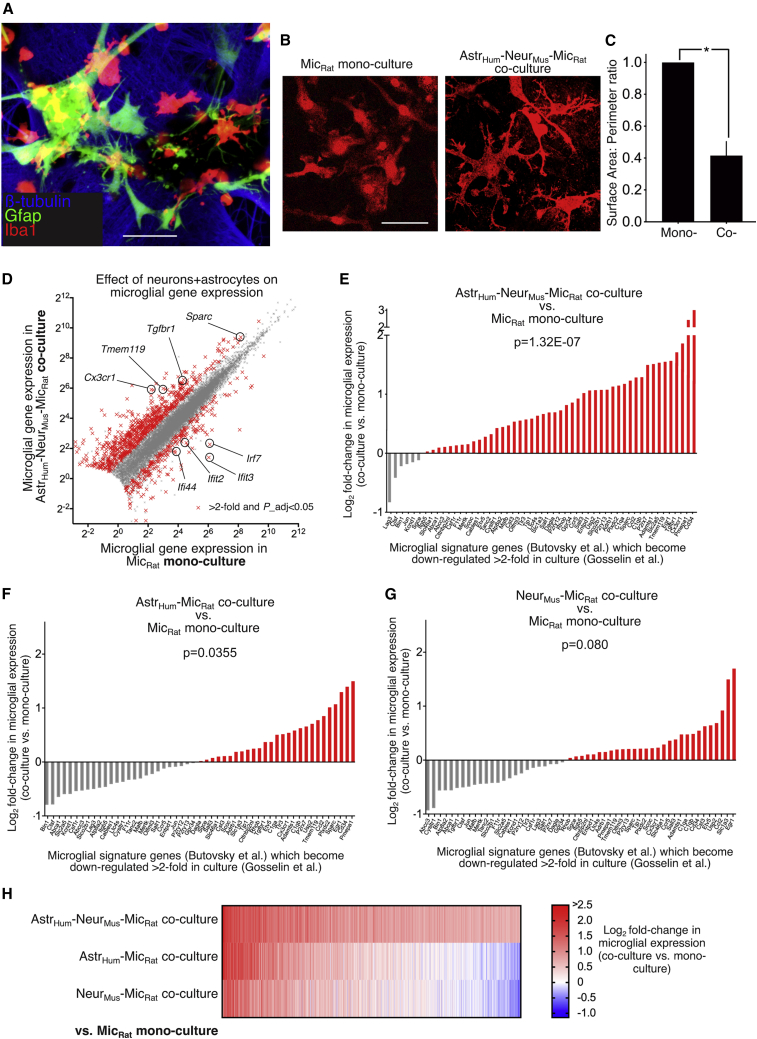
(A) A triple stain of the astrocyte/neuron/microglia co-culture with the indicated antibodies. Scale bar, 50 μm.
(B) Example images of Iba1-stained microglia in mono- or co-cultures as indicated. Scale bar, 20 μm.
(C) Surface area/perimeter ratio was calculated in mono- and co-culture. ∗p = 0.007 (t = 6.662, degrees of freedom [df] = 3), paired t test (120 cells analyzed per condition across n = 4 independent biological replicates).
(D) The influence of neuron-astrocyte co-culture on the microglial transcriptome. RNA-seq was performed on RNA extracted from (rat) microglia mono-cultures and (rat) microglia co-cultured with (human) astrocytes and (mouse) neurons. Both sets of reads were subjected to the same Sargasso workflow to identify unambiguously rat (microglial) reads and FPKM of all 13,406 genes shown that average >1 FPKM across the datasets is plotted for mono-culture (x axis) versus co-culture (y axis). Highlighted with red crosses are the 982 genes whose expression is significantly changed (DESeq2 P_adj < 0.05) by >2-fold (n = 7 mono-culture; n = 4 co-culture). “N” refers here and throughout as independent biological replicates derived from different culture material on different occasions. Upregulated genes highlighted are example MHSGs (Butovsky et al., 2014); downregulated genes highlighted are example interferon-related gene (IRG) cluster members (Friedman et al., 2018).
(E) Neurons and astrocytes combined boost microglial signature genes which become suppressed in vitro. Genes considered are those expressed >0.5 FPKM in our data and within the group of microglia signature genes defined by Butovsky et al. (2014), which are downregulated >2-fold after microglia were maintained in culture for 7 days compared to their expression immediately post-isolation from the intact brain, according to the data of Gosselin et al. (2017). For each gene, log2 fold change (log2FC) in microglial gene expression is shown relative to microglial mono-culture in (rat) microglia (mouse) neuron (human) astrocyte co-culture. The data were mined from the complete set shown in (D). ∗p = 1.3E-07, F (1,477) = 28.69 relates to main effect of co- versus monoculture condition on the gene set, two-way ANOVA.
(F and G) RNA-seq was performed on RNA extracted from (rat) microglia monocultures and (rat) microglia co-cultured with (human) astrocytes (F) or (mouse) neurons alone (G). log2FC in microglial gene expression is shown relative to microglial monoculture of the same genes as in (E). p = 0.036, F(1,424) = 4.45 (F), p = 0.080, F(1,424) = 3.09 (G), two-way ANOVA.
(H) Heatmap of the log2FC in microglial gene expression in the three different types of co-culture. 1,550 genes significantly induced >1.5-fold (DESeq2 P_adj < 0.05) are shown.