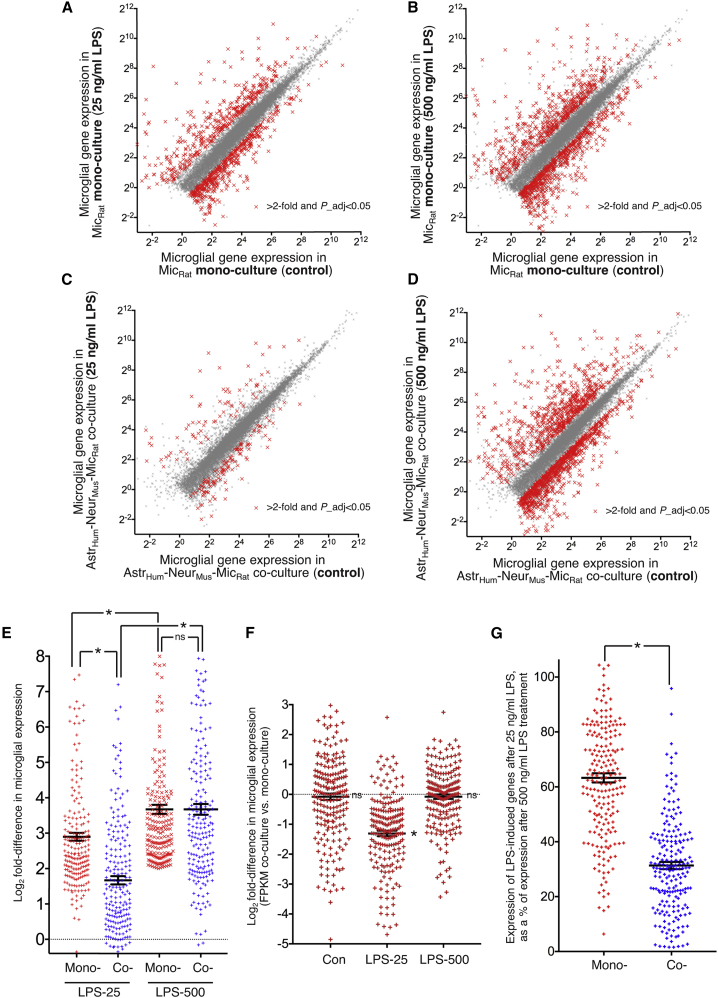
Figure 5.
Neurons and astrocytes regulate the microglial response to LPS
(A–D) LPS-induced microglial gene expression in microglial monoculture (A and B) and co-culture (C and D) in response to 16-h LPS treatment at 25 ng/mL (A and C) or 500 ng/mL (B and D). FPKM of all genes shown that average >1 FPKM across the datasets is plotted for control (x axis) versus LPS (y axis). Highlighted with red crosses are the genes whose expression is significantly changed (DESeq2 P_adj < 0.05) by >2-fold (n = 4).
(E) Log2FC in microglial gene expression is shown in the indicated cultures and in response to the indicated concentrations of LPS. The set of genes analyzed is the 206 induced >4-fold by the high does (500 ng/mL) of LPS in microglial monocultures is shown for all conditions. Mono-, microglial monoculture; Co-, (rat) microglia (mouse) neuron (human) astrocyte co-culture. ∗p values (left to right): <0.0001, 0.0001, <0.0001; ns, >0.9999; two-way ANOVA followed by Tukey’s post hoc test (n = 206).
(F) For the same set of the LPS-responsive genes in (E), we calculated the Log2 fold-change in microglial gene expression in co-culture versus monoculture, under the three different experimental treatments (con, 25 ng/ml LPS, 500 ng/ml LPS). p values (left to right):0.478 (ns), 3.05E-8 (∗), 0.141 (ns); 2-tailed paired Student’s t test on FPKM values in monoculture versus co-culture under each treatment condition.
(G) For the same set of the LPS-responsive genes in (E) and (F), the expression level (FPKM) of each gene under 25-ng/mL LPS conditions was calculated as a percentage of that gene’s expression level under 500-ng/mL LPS conditions. ∗p = 3.01E-39, unpaired two-tailed Student’s t test (n = 206)