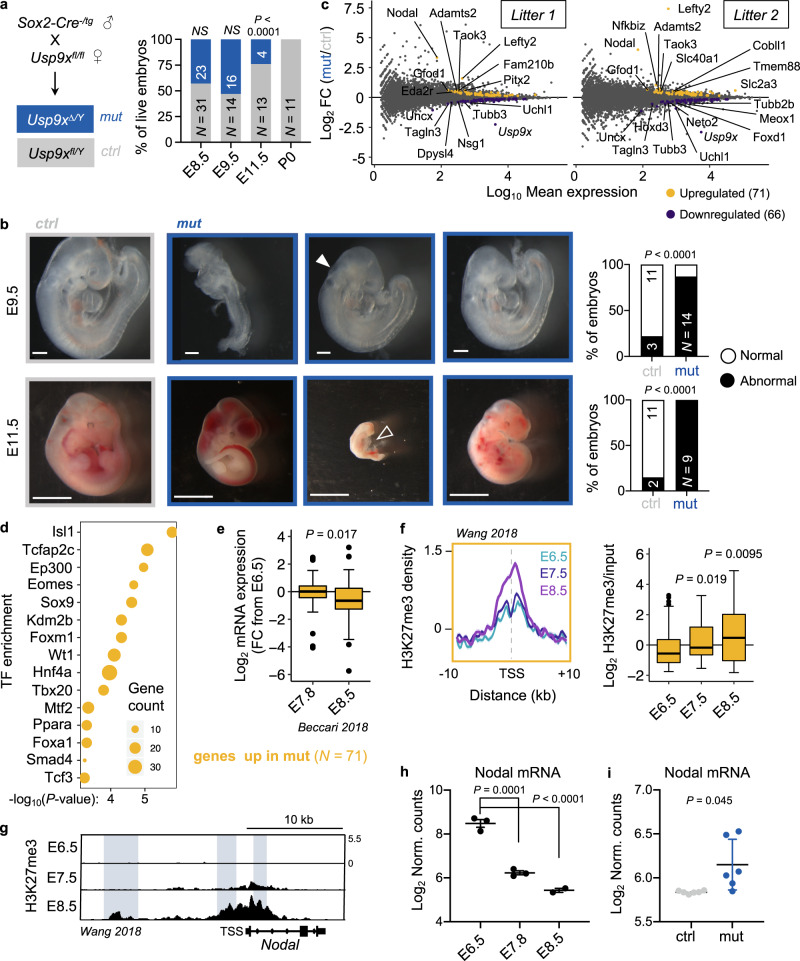
Fig. 2. Usp9x-mutant embryos arrest at E9.5–E11.5 and display defective repression of early lineage programs marked by H3K27me3.
a Genetic cross to delete Usp9x in epiblast derivatives of postimplantation embryos. Quantification of recovered (live) male embryos at several postimplantation stages (right). b Sample images and quantification of control and mutant embryo phenotypes. Relative to controls (left), E9.5 embryos show variable developmental delay, with closed arrow indicating an open anterior neuropore. E11.5 embryos show a range of phenotypes, from hemorrhage to severe delay and death (tally includes dead embryos). Open arrow indicates pericardial edema. Scale bars = 250 µm (E9.5), 2.8 mm (E11.5), with N indicated. c MA plots of expression changes by RNA-seq in two litters of Usp9x mutants versus controls (at E8.5). 3 mutants and 3 controls were sequenced per litter (N = 12 embryos total; see Supplementary Fig. 3b-d). d Enrichr analysis of the top-enriched transcription factors (TF) that bind to the genes upregulated in Usp9x-mutant embryos in various cell types. e Expression of the 71 genes upregulated in Usp9x mutants during wild-type development56. FC, Fold-change relative to E6.5 embryos. f Distribution and boxplot quantification of H3K27me3 levels59 over the promoters of genes upregulated in Usp9x mutants (10 kb upstream, 1 kb downstream of TSS). g Representative genome browser tracks of H3K27me3 in wild-type embryos (E6.5–E8.5) at the Nodal locus59. Known enhancer elements are highlighted and show gains of H3K27me3. h Nodal mRNA expression in wild-type development56. i Nodal mRNA expression in E8.5 Usp9x-mutant or control embryos. Boxplot hinges show the first and third quartiles, whiskers show ±1.5*IQR and center line shows median of 2–3 biological replicates (e, f). Data are representative of 2–3 biological replicates (g), mean ± s.e.m. of 2–3 biological replicates (h), or 6 biological replicates (i). P-values by χ2 test (a, b), Fisher’s exact test (d), two-tailed Student’s t-tests with Welch’s correction (e, i), two-tailed Wilcoxon rank-sum tests (f), and ANOVA with Dunnett’s multiple comparison test to E6.5 (h). χ2 = 19.78 (a), 85.19 (b, top), 147.8 (b, bottom).