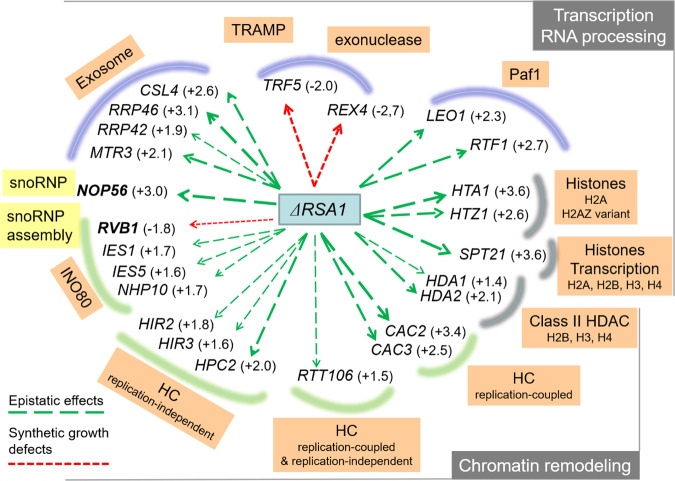
Fig. 1. Data from the GIM screen.
Two independent screens were performed, one with a mutant strain in which the RSA1 gene was deleted (rsa1Δ::NatR) and the other with a mutant used as reference strain. Two independent experiments were performed for each of these screens. The query strain and the reference strain were mated with a pool of strains containing all the viable strains from the haploid gene deletion collection. After selection of heterozygous diploids, sporulation and selection of the haploid, double mutants were grown for ~18 generations in rich liquid medium (YPD). Microarrays were used to measure the relative abundance of double mutants with query versus the reference populations. Normalized results are expressed as log2(Q/R) (see Methods section). Negative values indicate a synthetic growth defect. Positive values reveal either epistatic (buffering) or suppressive (alleviating) interactions between RSA1 and the selected genes. The genetic interactions with RSA1 are indicated by green arrows for the positive log2(Q/R) values and by red arrows for the negative values. The exact values are given in parentheses. HC heterochromatin.