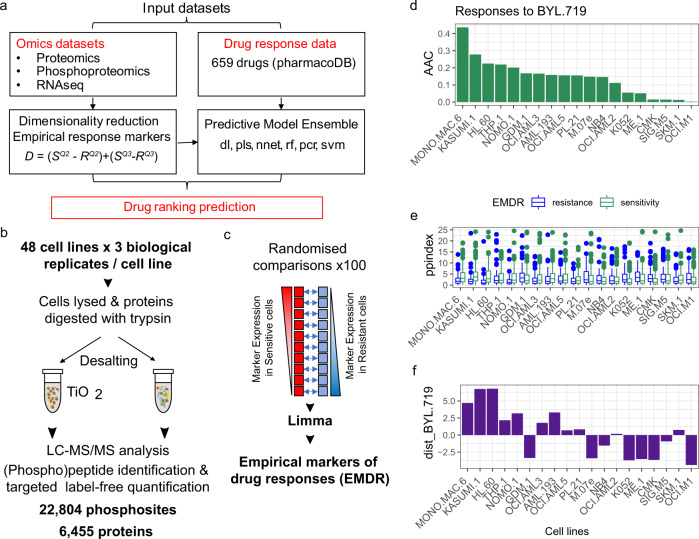
Fig. 1. Overview of Drug Ranking Using Machine Learning (DRUML).
a Drug response (AAC) values were modeled for 659 drugs with different DL/ML methods. Of these, 466 produced empirical markers of drug response and the responses for 411 drugs were reliably modeled by at least one learning algorithm. The input for DL/ML model generation are averaged values of empirical markers of drug responses (EMDRs), which are combined to derive a distance metric D. For each drug d and for each biological sample b, Dd,b = [(SQ2-RQ2) + (SQ3-RQ3)], where SQ2 and SQ3 are median and third quantile expression values of empirical markers increased in cells sensitive to a given drug, respectively; and RQ2 and RQ3 are median and third quartile expression values of empirical markers increased in cells resistant to the same drug. b LC-MS/MS workflow for the generation of proteomic and phosphoproteomic datasets used to train DRUML. c Approach to obtain empirical markers of drug responses (EMDRs). d Response values for BYL-719 obtained from PharmacoDB (n = 19). To determine empirical markers of drug sensitivity and resistance for BYL-719, cell lines are split into sensitive and resistant groups based on area above curve (AAC) values and Empirical Bayes Statistics of linear models were used to identify response markers by resampling. e Boxplot of the distributions of empirical markers of resistance and sensitivity in phosphoproteomics data acquired for the cell lines shown in panel d (measured in triplicate), boxplot with median center, interquartile box boundaries and range upper and lower hinges. f BYL-719 D values for the named cell lines calculated from the EMDR distributions shown in (e). Learning algorithms were random forest (rf), cubist, bayesian estimation of generalized linear models (bglm), partial least squares (pls), principal component regression (pcr), support vector machine (svm), deep learning (dl) and neural network (nnet).