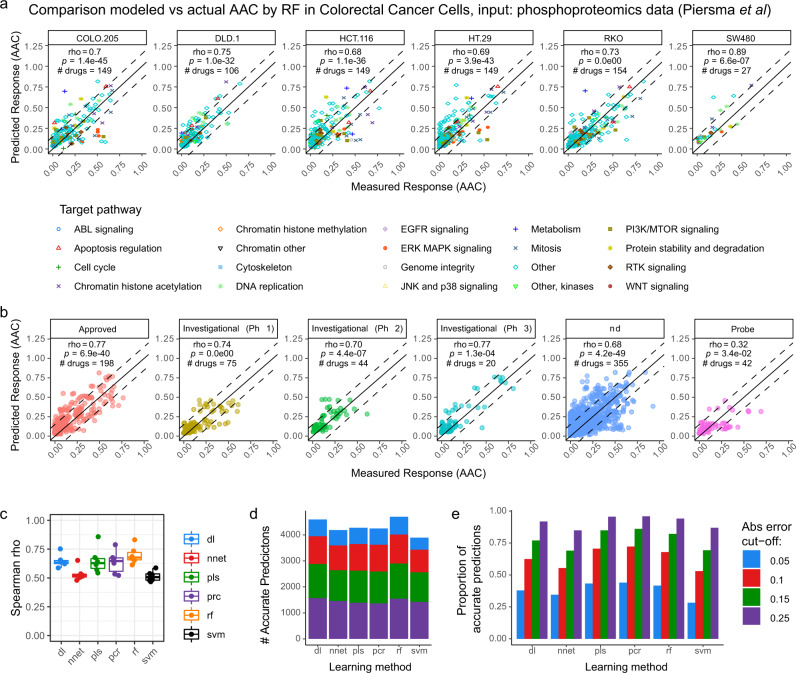
Fig. 5. Accuracy assessment of DRUML to rank drugs based on efficacy using an independent phosphoproteomics dataset.
DRUML was used to predict responses to 389 drugs in the colorectal cancer (CRC) cell lines shown using phosphoproteomic data obtained from Piersma et al (Jimenez lab, PRIDE PXD001550) a, b Scatter plots of measured and predicted drug responses. Dotted lines represent 10% absolute error margin, each data-point represents a drug prediction and statistical significance of drug ranking was assessed by measuring Spearman’s rank correlation coefficient. a Model performance across cell lines (n = 8). Drugs are colored by mode of action. b Comparison of measured and predicted drug responses binned by developmental stage of the drug. c Distribution of Spearman rho values by ML model. Boxplots represent median centers, interquartile box boundaries and range upper and lower hinges. d, e Number (d) and proportion (e) of accurate predictions of drugs for all cell lines within 0.05, 0.1, 0.15 and 0.25 absolute errors. Learning algorithms were random forest (rf), cubist, bayesian estimation of generalized linear models (bglm), partial least squares (pls), principal component regression (pcr), support vector machine (svm), deep learning (dl) and neural network (nnet).