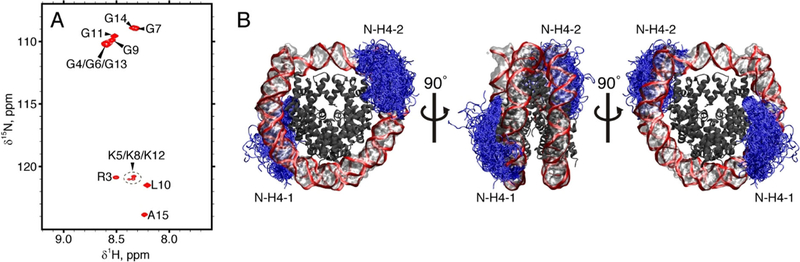
Figure 1.
(A) 1H,15N-HSQC spectrum of mononucleosomes containing 15N-labeled H4. (B) Dynamics of N-H4 tails (pictured as blue tubes) in mononucleosome according to the MD simulation data. To generate this plot, a 2-μs trajectory of NCP in TIP4P-D water was sampled with the step of 10 ns; the extracted frames were overlaid onto the reference structure 3LZ0. The DNA backbone is shown as a red band, bases and sugars are shown as a semi-transparent surface, the bodies of histone proteins are dark grey (with tails of H3, H2A, and H2B histones not shown). The visible asymmetry in spatial distribution of the N-H4–1 and N-H4–2 tails reflects a limited convergence of the simulation.