Fig. 2. Structure of the ROQ1 LRR and C-JID (PL domain) binding to XopQ.
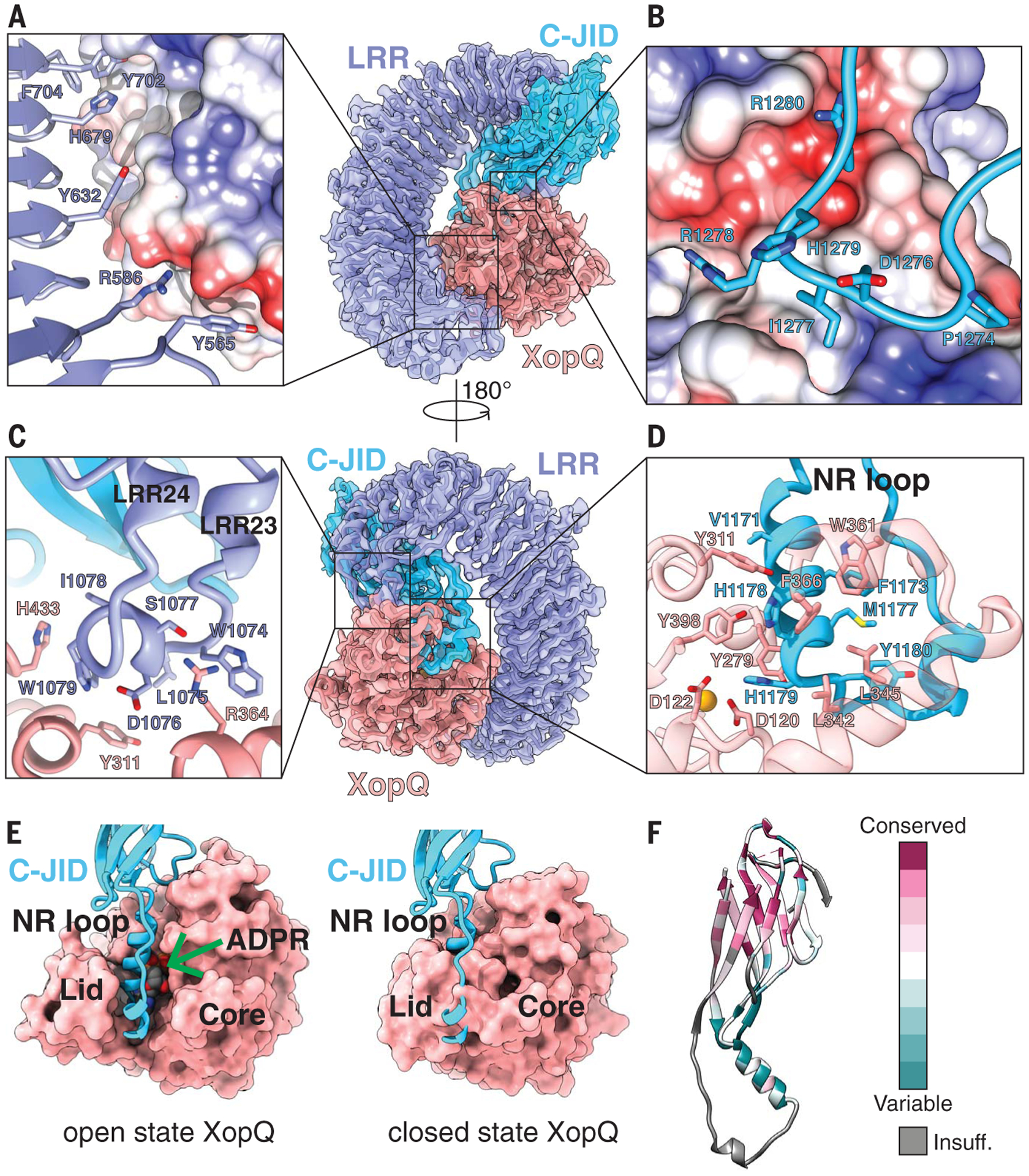
(A) Surface contacts between the N-terminal region of the LRR, shown with a violet ribbon, and XopQ, represented by its Coulombic surface potential. (B) Surface contacts made by the loop between β-strands 7 and 8 of the C-JID domain (light blue) and XopQ. (C) The elongated LRR between repeats 23 and 24 (violet) interacting with XopQ (salmon). (D) Interactions between the NR loop (light blue) and active-site residues of XopQ required for ADPR binding. Catalytic Ca2+ is shown in gold. (E) Left: Structure of XopQ in the open conformation built from our cryo-EM density, with the NR loop inserted into the active-site cleft. The position of ADPR (green arrow) from the close state of XopQ (PDB: 4P5F) is modeled to show its overlapping position with the NR loop. Right: ADPR-bound, closed state of XopQ. The NR loop is modeled to demonstrate the clashes that would occur upon XopQ closure. (F) Residue conservation of the C-JID. Regions where too few sequences aligned to calculate a reliable conservation score are colored in gray (labeled “Insuff.”).
