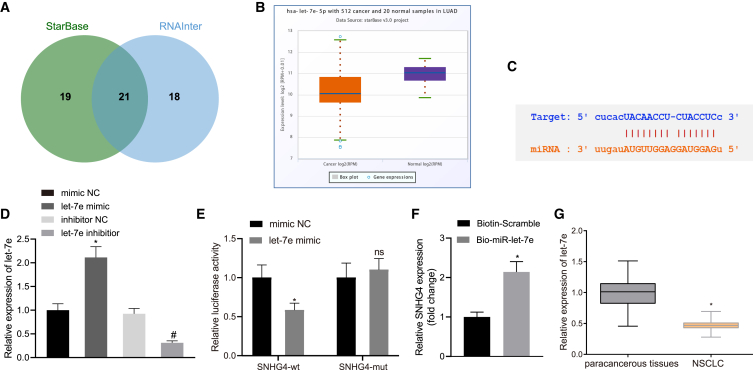
Figure 2.
lncRNA SNHG4 binds with miR-let-7e
(A) Common 21 putative miRNAs binding with lncRNA SNHG4 stood out between two public databases, i.e., starBase (http://starbase.sysu.edu.cn/) and RNAInter (http://www.rna-society.org/rnainter/), shown by the Venn diagram. (B) Expression box of miR-let-7e between lung cancer tissues and normal tissues in the starBase database. (C) Putative miR-let-7e binding sites in the 3′ UTR of lncRNA SNHG4 by the starBase database. (D) The expression of miR-let-7e was determined by qRT-PCR in H1299 cells treated with miR-let-7e mimic and miR-let-7e inhibitor. (E) Luciferase activity at the promoter of the reporter gene containing the seed sequence in the 3′ UTR of lncRNA SNHG4 and, accordingly, the mutant reporter gene. (F) RNA pull-down of SNHG4 by miR-let-7e detected by qRT-PCR. (G) The expression of miR-let-7e was determined between NSCLC tissues (n = 50) and matched paracancerous lung tissues (n = 50) by qRT-PCR. In (D) and (E), ∗p < 0.05 (compared with H1299 cells treated with mimic NC) and #p < 0.05 (compared with H1299 cells treated with inhibitor NC) by unpaired t test. In (F), ∗p < 0.05 compared with biotin-scramble by an unpaired t test. In (G), ∗p < 0.05 compared with matched noncancerous lung tissues by a paired t test.