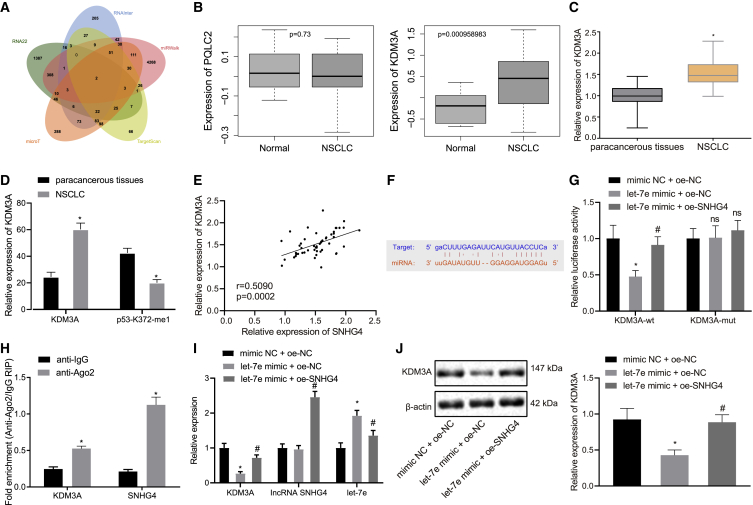
Figure 4.
lncRNA SNHG4 bound with miR-let-7e and upregulated KDM3A
(A) Putative target genes of miR-let-7e among the RNA22 (https://cm.jefferson.edu/rna22/), RNAInter (http://www.rna-society.org/rnainter/), miRWalk (http://mirwalk.umm.uni-heidelberg.de/), TargetScan (http://www.targetscan.org/vert_72/), and microT (http://diana.imis.athena-innovation.gr/DianaTools/index.php?r=microT_CDS/index) databases. (B) Expression box of PQLC2 and KDM3A between lung cancer tissues and normal tissues in the starBase database. (C) The expression of KDM3A was determined by qRT-PCR in cancerous tissues (n = 50) and matched noncancerous lung tissues (n = 50). (D) Immunohistochemical staining for KDM3A and p53-k372me1 in cancerous tissues (n = 50) and matched noncancerous lung tissues (n = 50). (E) Pearson correlation analysis of KDM3A and lncRNA SNHG4. (F) Putative miR-let-7e binding sites in the 3′ UTR of KDM3A by the starBase database. (G) Luciferase activity at the promoter of the reporter gene containing the seed sequence in the 3′ UTR of KDM3A and, accordingly, the mutant reporter gene in the presence of miR-let-7e mimic and/or expression vector containing the lncRNA SNHG4. (H) Anti-Ago2 RIP in H1299 cells transiently overexpressing lncRNA SNHG4. (I) The expression of KDM3A was determined by qRT-PCR in H1299 cells. (J) Western blots and quantification of KDM3A in H1299 cells, normalized to β-actin expression. ∗p < 0.05 (compared with H1299 cells treated with oe-NC with or without mimic NC) and #p < 0.05 (compared with H1299 cells treated with miR-let-7e mimic with oe-NC) by unpaired t test or Tukey’s test-corrected one-way ANOVA. In (C) and (D), ∗p < 0.05 compared with matched noncancerous lung tissues by paired t test.