Figure 3.
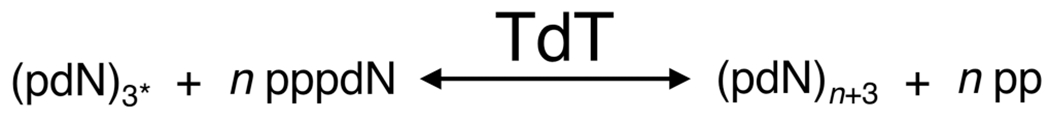
TdT polymerization of ssDNA. Here, a 3′-hydroxylated initiator strand (also termed an acceptor) is required [in the presence of a divalent cation (e.g., Mg2+)] for TdT to catalyze ssDNA using a deoxyribonucleoside triphosphate monomer (dNTP, also termed a donor). Here dNTP (pppdN) monomers are the substrate for addition of TdT to the 3′ end of an initiator strand (pdN)3* to generate ssDNA (pdN)n+3 with the release of pyrophosphate (pp). 3* indicates the length must be at least 3 nt. There are two primary isoforms of the TdT gene, TdTL (3′ exonuclease activity only, arrow right to left) and TdTS (3′ terminal transferase only, arrow left to right).
