Figure 6.
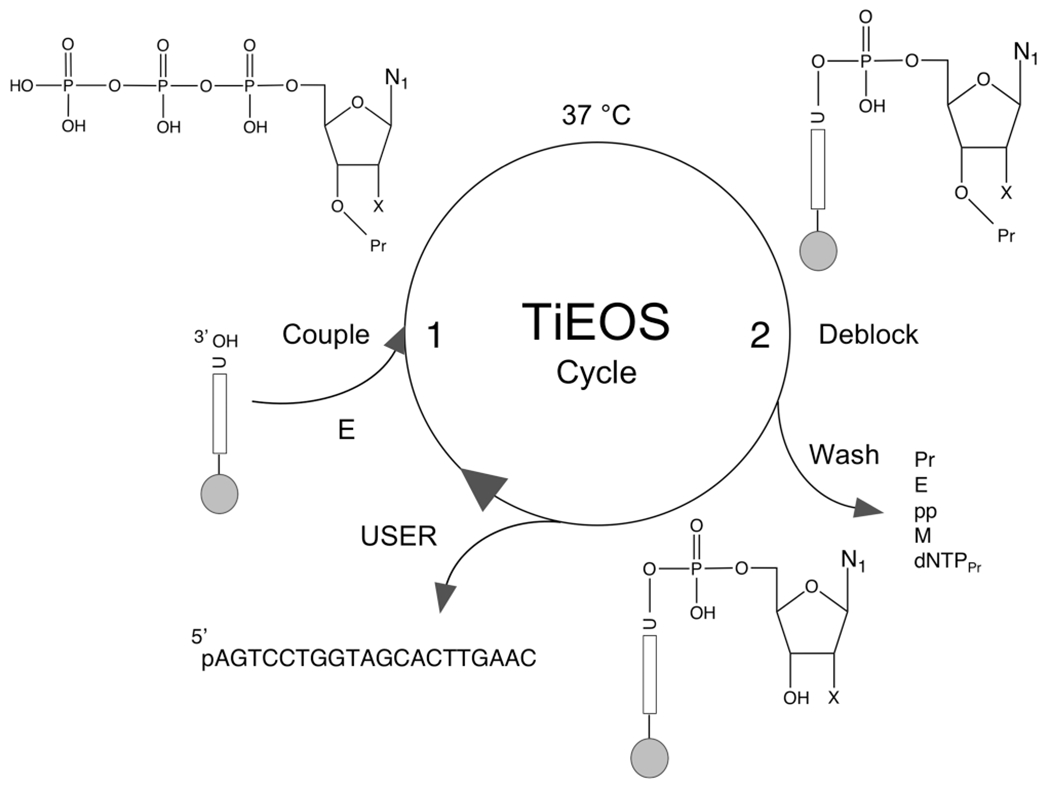
Proposed cycle for template-independent enzymatic oligonucleotide synthesis (TiEOS). In step 1, an incoming 3′-protected (Pr) dNTPPr (or NTPPr) is coupled to a 20 nt initiator (e.g., 37 °C, 30 s), which allows for enzyme (TdT) attachment and polymerization. Here the initiator is chemically presynthesized and covalently tethered {e.g., carbodiimide chemistry to a solid substrate [e.g., superparamagnetic beads (SPMB), silicon, or glass]}. A 20 nt long initiator was chosen to prevent steric hindrance at the surface of the beads during polymerization. In step 2, newly added dNTPPr is deblocked at the 3′ to hydroxyl (dNTP), followed by a wash step to remove protecting groups, enzyme, pyrophosphate, metal ions, and unincorporated dNTPPr. The cycle is repeated until the FLP is completed. For the purposes of enzymatic target strand release, uracil is placed at the 3′ end of the initiator for a point of cleavage; uracil DNA glycosylase and endonuclease VIII (USER) cleave the target strand [now 5′-phosphorylated (p)] from the support.47 Legend: gray spheres, solid support; vertical bars, initiator strands (>20 nt); E, enzyme (e.g., terminal transferase); U, uridine; N, nucleobase (see Figure 1B); p, phosphate; O, oxygen; pp, pyrophosphate; M, metal (divalent cation); X, either H (dNTP), OH (NTP for RNA synthesis), or a protecting group (Figure 8).
