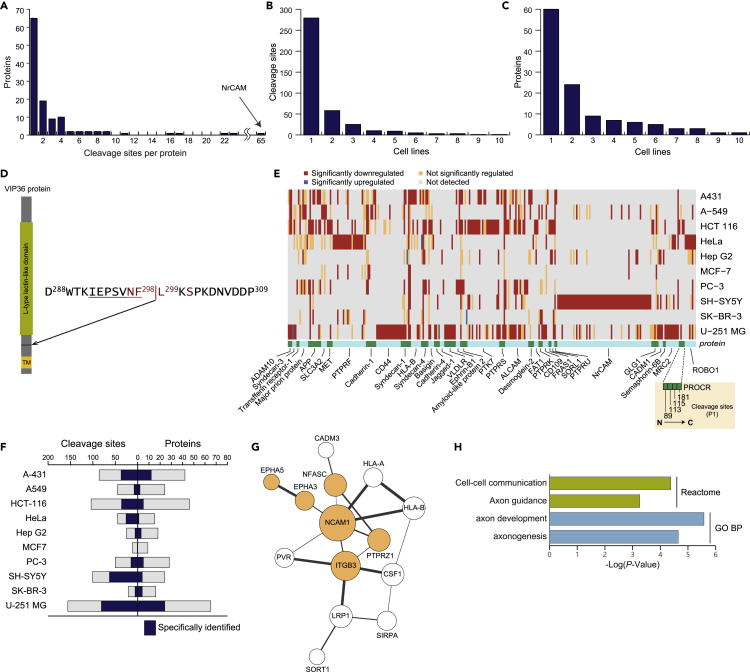
Figure 5.
Cell-type-specificity of shedding
(A) The distribution of the number of cleavage sites per protein is depicted for single-pass type-I and type-II transmembrane proteins and GPI-anchored proteins.
(B and C) Histograms depicting the number of shedding substrate identified in 1–10 cell lines at the cleavage site level (B) and at the protein level (C).
(D) VIP36 cleavage site. The amino acids that were previously suggested to be essential for cleavage are highlighted in red (Shirakabe et al., 2011). The sequence of the peptide identified in this study is underlined. TM, transmembrane domain.
(E) Heatmap depicting membrane proteins shed by cleavage at differential sites in different cell lines (36 proteins). Each gray, yellow, red, or blue box designates a cleavage site. Cleavage sites are arranged in the order from N- to C-terminus from left to right in each protein, as the example of PROCR shows. At the bottom, to visualize the boundaries of proteins, green and light-blue boxes are alternately arranged in alphabetical order of UniProt accession from left to right.
(F) Distribution of the number of metalloprotease-regulated cleavage sites in the respective cell lines. The number of cleavage sites specifically identified in each cell type is highlighted in color.
(G and H) Protein interaction network of shedding substrate proteins specifically identified in U-251 MG cells (G). Proteins without any interactions are excluded. Significantly enriched reactome and GO terms of interest are shown with the Benjamini-adjusted p values (H), and the proteins annotated with these terms are highlighted in color (F). All analyses were performed by STRING (v11). The network was visualized using Cytoscape (v3.8.0). Node size reflects the number of connections (direct edges), and edge line width reflects the combined score calculated by STRING.