Abstract
Gryllodes sigillatus is a cricket widely distributed throughout the world. In this study, we reported the first complete mitogenome sequence of Genus Gryllodes and inferred its phylogeny. The mitogenome of G. sigillatus was 16,369 bp and consisted of a control region and a typical set of 37 genes. It was AT-rich with strong codon usage bias and possessed a gene arrangement of trnE-trnS1-trnN. Phylogenetic analysis indicated G. sigillatus was sister species to Velarifictorus hemelytrus, together belonging to the Family Gryllidae. Our findings would contribute to understanding mitogenomic evolution and phylogeny of Ensifera.
Keywords: Gryllodes sigillatus, Gryllidae, mitochondrial genome, Phylogeny
Gryllodes sigillatus (Walker), a field cricket, also known as tropical house cricket, belongs to the Family Gryllidae with a wide distribution throughout the world (Otte 2006). It is a pest but sometimes also kept as a pet or animal feed in addition to a promising protein source for human diet (Ma et al. 2019; Daniso et al. 2020). G. sigillatus was first described in 1869 and then classified into different genera many times (Otte 2006). Insect mitogenome sequences have been extensively used to infer the evolution and phylogeny of metazoan at both deep and shallow taxonomic levels due to its fast mutation rate, conservation among conspecifics and lack of recombination (Cameron 2014; Li et al. 2019; Shaoli et al. 2018). Here, we reported the complete mitogenome sequence of G. sigillatus, the first in the Genus Gryllodes, which would contribute to evolution and phylogeny of Ensifera.
The samples of G. sigillatus were collected in the wild grass field from Nankang Town (21°34′41.08″N, 109°24′57.60″E), Beihai, Guangxi Province, China, and stored in the Medical Biology Institute, Wannan Medical College (voucher YGS201001). The species was identified by morphological characteristics and sequence analysis of cox1 and cytb. Genomic DNA was extracted from muscle tissue using a TIANamp Genomic DNA Kit (TIANGEN, Beijing, China) and sequenced by Illumina NovaSeq 6000 platform (Illumina, San Diego, CA, USA). The mitogenome sequence was assembled by GetOrganelle v1.7.0 (Jin et al. 2020), checked manually with Geneious 10.2.2 (Kearse et al. 2012) and annotated using MitoZ version 2.3 (Meng et al. 2019).
The complete mitogenome of G. sigillatus was a circular DNA molecule of 16,369 bp in length. It comprised of a control region (also called AT-rich or D-loop region) and a typical set of 37 genes coding for 13 proteins (PCGs), two ribosomal RNA and 22 transfer RNA. Similar to mitogenomes of other Gryllidae species (Ma and Li 2018; Ma et al. 2019), G. sigillatus possessed a unique gene arrangement of trnE-trnS1-trnN. Twenty genes were encoded on the majority strand (J-strand). The overall AT content was 70.40%. AT- and GC-skew were 0.07 and −0.31, respectively, indicating that the mitogenome of G. sigillatus was AT-rich and adenine was preferred. For PCGs, all started with ATN except the cox1 with TCG as the start codon, and terminated with TAA, TAG or T. The AT percentage of 80.17% at the third codon position was higher than that at other two positions. Moreover, Leu was the most abundant (15.53%) among the used 20 amino acids and codon usage biases were strong with range of relative synonymous codon usage from 0.00 (AGG) to 3.22 (TTA).
To further explore the phylogeny of Grylloidea, we reconstructed the phylogenetic trees based on PCGs sequences of released mitogenomes in GenBank using the maximum likelihood (ML), maximum parsimony (MP) and Bayesian inference (BI) methods that were performed by RaxML GUI 2.0 (Silvestro and Michalak 2012), PAUP* v4.0a168 (Swofford 2003) and MrBayes 3.2.6 (Ronquist et al. 2012), respectively. The substitution saturation was evaluated with DAMBE 7.0 (Xia 2018), and the most suitable models for each of PCGs were assessed by Modeltest-NG v0.1.6 (Darriba et al. 2020).
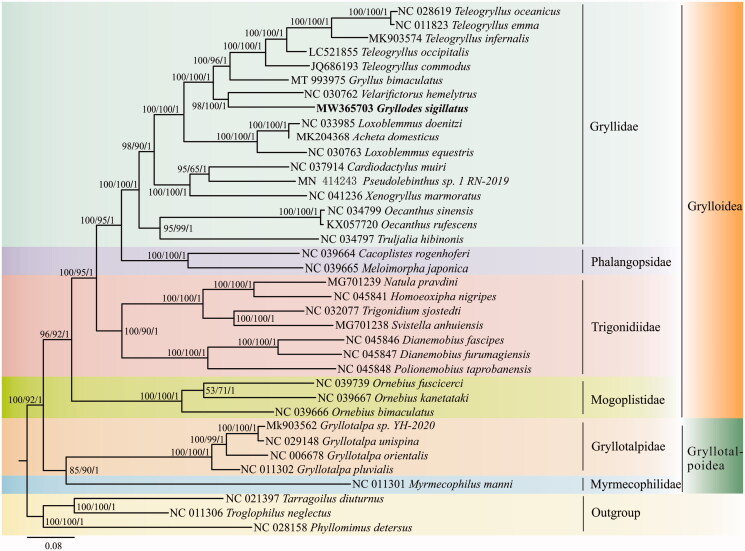
The topology of ML, MP and BI trees were highly consistent only with slightly difference for support values at some nodes (Figure 1). The results indicated that G. sigillatus was sister species to Velarifictorus hemelytrus and belonged to Gryllinae subfamily in the Family Gryllidae. Oecanthus and Truljalia genera grouped together at the base of Gryllidae. It was identical with the result of Ma et al. (Ma et al. 2019). Moreover, the Family Gryllidae, Phalangopsidae and Trigonidiidae clustered a monophyly sharing a gene arrangement of trnE-trnS1-trnN, whereas Gryllotalpoidea was at the base of Grylloidea with a trnN-trnS1-trnE gene arrangement.
Figure 1.
Phylogenetic tree of 37 Ensifera species based on 13 PCGs sequences from mitogenomes and inferred with maximum likelihood (ML), maximum parsimony (MP) and Bayesian inference (BI) methods, respectively. Of them, three mitogenomes NC021397, NC011306 and NC028158 are selected as outgroups. Bootstrap/posterior probability values are displayed on the branches in the order ML/MP/BI, and values less than 50/0.5 are not shown. GenBank accession numbers are listed in front of species name and bold text represents the species in this study.
Funding Statement
This study was supported by the Key Scientific Research Project of Wannan Medical College (Grant No. WK2020Z08) and College students Innovation and Enterprise Training Foundation of Anhui Province (Grant No. s201910368003 and s201910368016).
Data availability statement
The mitogenom sequence data are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW365703. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA693646, SAMN17393255, and SRR13495188, respectively.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- Cameron SL. 2014. Insect mitochondrial genomics: implications for evolution and phylogeny. Annu Rev Entomol. 59:95–117. [DOI] [PubMed] [Google Scholar]
- Daniso E, Tulli F, Cardinaletti G, Cerri R, Tibaldi E.. 2020. Molecular approach for insect detection in feed and food: the case of Gryllodes sigillatus. Eur Food Res Technol. 246(12):2373–2381. [Google Scholar]
- Darriba D, Posada D, Kozlov AM, Stamatakis A, Morel B, Flouri T.. 2020. ModelTest-NG: A New and Scalable Tool for the Selection of DNA and Protein Evolutionary Models. Mol Biol Evol. 37(1):291–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin J, Yu W, Yang J, Song Y, dePamphilis CW, Yi T, Li D.. 2020. GetOrganelle: a fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21(1):241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, et al. . 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 28(12):1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Chen Q, Wen M, Wang J, Wang Y, Ren B.. 2019. Phylogeny and acoustic signal evolution of a pure tone song katydid Pseudophyllus titan (Orthoptera: Tettigoniidae) based on the complete mitogenome. Mitochondrial DNA A DNA Mapp Seq Anal. 30(3):385–396. [DOI] [PubMed] [Google Scholar]
- Ma C, Li J.. 2018. Comparative analysis of mitochondrial genomes of the superfamily Grylloidea (Insecta, Orthoptera) reveals phylogenetic distribution of gene rearrangements. Int J Biol Macromol. 120(Pt A):1048–1054. [DOI] [PubMed] [Google Scholar]
- Ma C, Wang Y, Zhang L, Li J.. 2019. Mitochondrial genome characterization of the family Trigonidiidae (Orthoptera) reveals novel structural features and nad1 transcript ends. Sci Rep. 9(1):19092–19092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng G, Li Y, Yang C, Liu S.. 2019. MitoZ: a toolkit for animal mitochondrial genome assembly, annotation and visualization. Nucleic Acids Res. 47(11):e63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otte D. 2006. Gryllodes sigillatus (Walker) is a valid species distinct from Gryllodes supplicans (Walker). T Am Entomol Soc. 132(1):223–227. [Google Scholar]
- Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP.. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 61(3):539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaoli M, Hao Y, Chao L, Yafu Z, Fuming S, Yuchao W.. 2018. The complete mitochondrial genome of Xizicus (Haploxizicus) maculatus revealed by Next-Generation Sequencing and phylogenetic implication (Orthoptera, Meconematinae). ZK. (773):57–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestro D, Michalak I.. 2012. raxmlGUI: a graphical front-end for RAxML. Org Divers Evol. 12(4):335–337. [Google Scholar]
- Swofford DL. 2003. PAUP*: phylogenetic analysis using parsimony (*and other Methods) Version 4. Sinauer, Sunderland, Massachusetts, USA. Nat Biotechnol. 18:233–234. [Google Scholar]
- Xia X. 2018. DAMBE7: new and improved tools for data analysis in molecular biology and evolution. Mol Biol Evol. 35(6):1550–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The mitogenom sequence data are openly available in GenBank of NCBI at (https://www.ncbi.nlm.nih.gov/) under the accession no. MW365703. The associated BioProject, Bio-Sample, and SRA numbers are PRJNA693646, SAMN17393255, and SRR13495188, respectively.