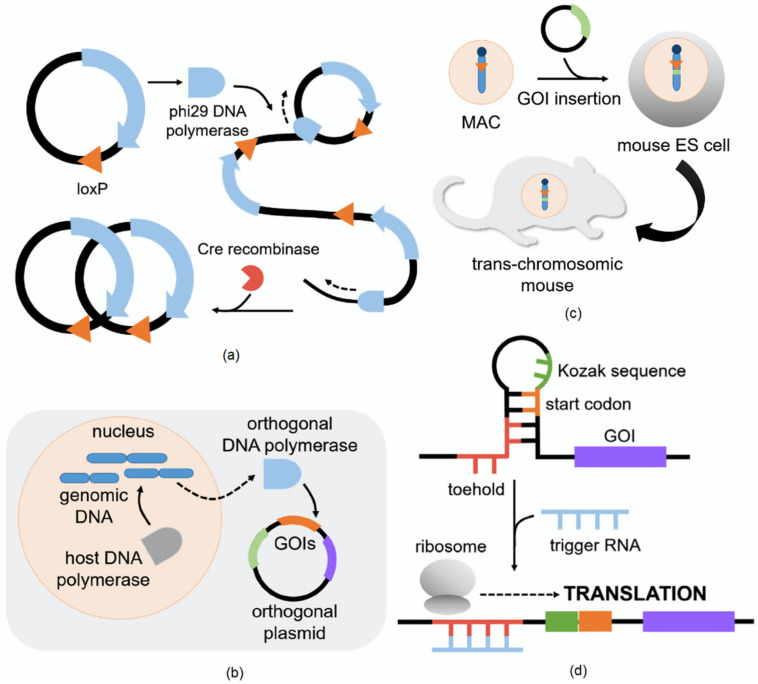
Figure 1.
Synthetic biology techniques to improve orthogonality of expression vectors introduced into animal systems. (a) Minimal TTcDR [59]. A plasmid which encodes its own DNA polymerase, phi29, replicates via rolling circle replication. Cre recombinase cuts at the loxP sites, which results in recircularization of the DNA in vitro. (b) OrthoRep system in yeast [60]. Native machinery from yeast generates an orthogonal DNA polymerase that solely replicates the desired plasmid. This separates the replication of the genes of interest (GOIs) in the cytosol from nuclear replication of endogenous genes. While this has not yet been tested in animal systems, the yeast system acts as a proof of concept for compartmentalization of DNA replication between the nucleus and cytoplasm in eukaryotes. (c) Mouse artificial chromosome (MAC) stably expressed in mouse embryonic stem (ES) cells [61]. Takiguchi et al. constructed the MAC vector from a truncated native mouse chromosome. The incorporation of a Cre-loxP-mediated platform enables insertions of a GOI, and the modified MAC can be transferred into mouse ES cells to form trans-chromosomic mice. (d) Mammalian toehold switch for a post-transcriptional circuit [62]. Dependent on RNA–RNA interactions, it contains a toehold sequence complementary to a trigger RNA, a stem–loop structure that includes a Kozak sequence, and a start codon upstream of the gene of interest. The translation of the gene is constitutively repressed. Once a trigger RNA binds to the toehold sequence, the stem–loop structure is disrupted and exposes the Kozak sequence and start codon in order to initiate translation.