A near-complete genome sequence was obtained for a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VOC) 202012/01 strain obtained from an oropharyngeal swab sample from a Peruvian patient with coronavirus syndrome who had contact with an individual who had recently returned from England.
ABSTRACT
A near-complete genome sequence was obtained for a novel severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variant of concern (VOC) 202012/01 strain obtained from an oropharyngeal swab sample from a Peruvian patient with coronavirus syndrome who had contact with an individual who had recently returned from England.
ANNOUNCEMENT
The severe acute respiratory syndrome (SARS) that currently affects the world is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which belongs to the Betacoronavirus genus of the family Coronaviridae (1, 2). An article has recently been prepublished showing that the new variant designated variant of concern (VOC) 202012/01 has a greater transmission capacity, but it has not been shown to be more pathogenic (3).
During surveillance, we detected a female patient with coronavirus syndrome who claimed to have been in contact with a relative who had recently returned from England. This patient was confirmed as being infected with SARS-CoV-2 by quantitative reverse transcription (RT)-PCR using the Xpert Xpress SARS-CoV-2 test on the GeneXpert system (threshold cycle [CT] values for the E gene and N2 were 17.9 and 19.6, respectively). Viral RNA was purified from 200 μl of viral transport medium using the Maxwell 16 viral total nucleic acid purification kit in a Maxwell 16 instrument (Promega, Madison, WI, USA) following the manufacturer’s instructions. The purified RNA samples were processed by RT-PCR to amplify overlapping fragments, with primers designed using the SARS-CoV-2 genome from a Wuhan isolate, as reported previously (4). This study was approved by the institutional ethics committee at the Peruvian National Institute of Health (project code OI-045-20).
The amplification process used the SuperScript IV One-Step RT-PCR high-fidelity system kit (Invitrogen), and the amplified products were purified with the PureLink PCR purification kit (Invitrogen). The amplified fragments (2 ng) were processed using the Nextera XT DNA library preparation kit, index adapters, and the MiSeq sequencer (Illumina) following the procedure recommended by the manufacturer. The fastq files generated were processed in the Galaxy platform (5) and cleaned using Groomer v1.1.5 and Trimmomatic v0.38.0 (https://usegalaxy.org). Then, the genomes were assembled using SPAdes v3.12.0 and compared to the reference genome using CONTIGuator v2.7.4 (http://contiguator.sourceforge.net). Nucleotide and amino acid variations were detected using Geneious Prime software v2021.0.3. To perform the phylogenetic analysis, other relevant genomes were downloaded from the GISAID database, the genomes were aligned with MAFFT v7 (https://mafft.cbrc.jp/alignment/server), and a phylogenetic tree was built using the maximum likelihood algorithm (based on the GTR model and the gamma distribution) with the program MEGA X v7.0.26 (https://www.megasoftware.net). Default parameters were used for all software unless otherwise specified.
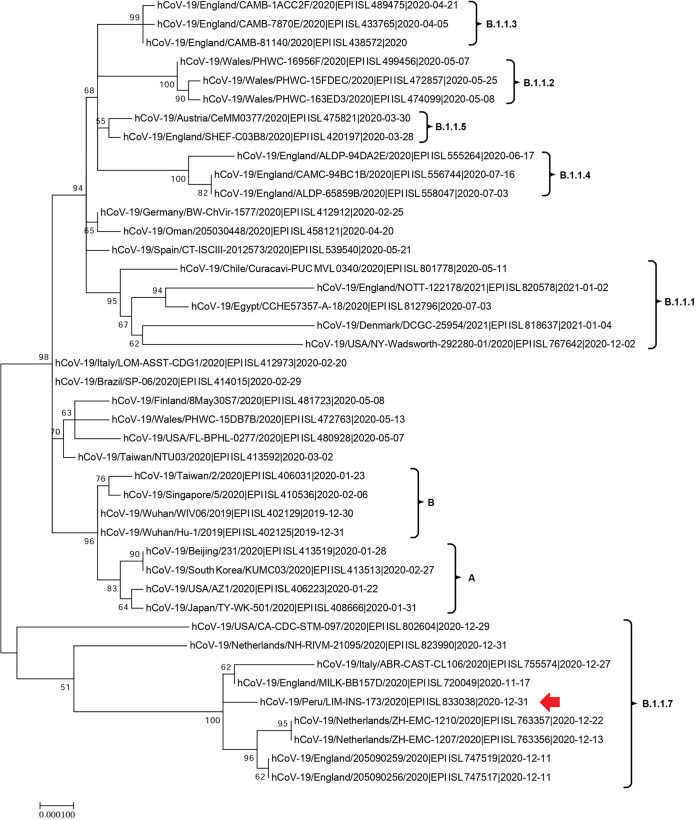
The genome of this viral isolate has two deletions (aminoacidic position 69 to 70 and position 144 to 145 deletions) and eight mutations (V483I, N501Y, A570D, D614G, P681H, T716I, S982A, and D1118H) in the S gene, similar to VOC 202012/01. Also, it presents the C241T mutation in the 5′ untranslated region, and it presents five nonsynonymous mutations (L730F, T1001I, A1708D, I2230T, and P4715L) and a deletion of three amino acids (S3575, G3676, and F3677) in the orf1ab gene. The C27972T mutation, which generates a premature stop, is present in open reading frame 8 (ORF8), while the N gene presents the D3L, D203K, G204R, and S235F variations. The lineage of this sample corresponds to B.1.1.7 according to the Pangolin system, and phylogenetic analysis indicates that it is homologous to the VOC 202012/01 isolates reported in the GISAID database (Fig. 1). This is the first report of this variant in Peru, and a study of the contacts of this patient is being carried out to evaluate how this new variant is spreading.
FIG 1.
Phylogenetic tree of the VOC 202012/01 strain detected in Peru (indicated by the red arrow) with relevant genomes (the GISAID accession number is indicated in each name). This tree was built with the maximum likelihood algorithm (GTR+G model) using MEGA X.
Data availability.
The sequence of the SARS-CoV-2 variant was deposited in the NCBI database (GenBank accession number MW494424.1) and the GISAID database (https://www.gisaid.org) (accession number EPI_ISL_833038). The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (accession number PRJNA623683).
ACKNOWLEDGMENTS
This work is part of the National Molecular Surveillance of SARS-CoV-2 in Peru and has been funded by the National Institute of Health of Peru.
We thank the team in charge of COVID-19 molecular diagnosis at the National Institute of Health of Peru for all of their efforts and contributions in the confirmation of cases. We also thank all of the personnel in charge of the surveillance and follow-up monitoring of cases and the professionals who allowed us to manage and obtain the supplies for this study.
REFERENCES
- 1.Wu F, Zhao S, Yu B, Chen Y-M, Wang W, Song Z-G, Hu Y, Tao Z-W, Tian J-H, Pei Y-Y, Yuan M-L, Zhang Y-L, Dai F-H, Liu Y, Wang Q-M, Zheng J-J, Xu L, Holmes EC, Zhang Y-Z. 2020. A new coronavirus associated with human respiratory disease in China. Nature 579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, Zhao X, Huang B, Shi W, Lu R, Niu P, Zhan F, Ma X, Wang D, Xu W, Wu G, Gao GF, Tan W, China Novel Coronavirus Investigating and Research Team . 2020. A novel coronavirus from patients with pneumonia in China, 2019. N Engl J Med 382:727–733. doi: 10.1056/NEJMoa2001017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Volz E, Mishra S, Chand M, Barrett JC, Johnson R, Geidelberg L, Hinsley WR, Laydon DJ, Dabrera G, O’Toole A, Amato R, Ragonnet-Cronin M, Harrison I, Jackson B, Ariani CV, Boyd O, Loman NJ, McCrone JT, Gonçalves S, Jorgensen D, Myers R, Hill V, Jackson DK, Gaythorpe K, Groves N, Sillitoe J, Kwiatkowski DP, COVID-19 Genomics UK (COG-UK) Consortium, Flaxman S, Ratmann O, Bhatt S, Hopkins S, Gandy A, Rambaut A, Ferguson NM. 2020. Transmission of SARS-CoV-2 lineage B.1.1.7 in England: insights from linking epidemiological and genetic data. medRxiv 2020.12.30.20249034. doi: 10.1101/2020.12.30.20249034. [DOI] [Google Scholar]
- 4.Padilla-Rojas C, Vega-Chozo K, Galarza-Perez M, Bailon Calderon H, Lope-Pari P, Balbuena- Torres J, Garcia Neyra D, Huaringa-Nuñez M, Rojas-Serrano N, Caceres-Rey O. 2020. Genomic analysis reveals local transmission of SARS-CoV-2 in early pandemic phase in Peru. bioRxiv 2020.09.05.284604. doi: 10.1101/2020.09.05.284604. [DOI]
- 5.Jalili V, Afgan E, Gu Q, Clements D, Blankenberg D, Goecks J, Taylor J, Nekrutenko A. 2020. The Galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2020 update. Nucleic Acids Res 48:W395–W402. doi: 10.1093/nar/gkaa434. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The sequence of the SARS-CoV-2 variant was deposited in the NCBI database (GenBank accession number MW494424.1) and the GISAID database (https://www.gisaid.org) (accession number EPI_ISL_833038). The raw reads were deposited in the NCBI Sequence Read Archive (SRA) database (accession number PRJNA623683).