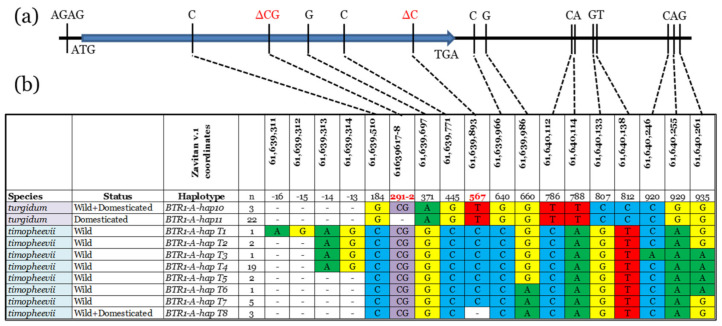
Figure 1.
(a) Schematic showing the locations of the sequence polymorphisms used in the BTR1-A haplotype analysis. Highlighted in red font are two frameshift deletions in the BTR1-A coding sequence. (b) Among the materials included in this study, a total of 10 BTR1-A haplotypes were identified, two within the T. turgidum species and eight within the T. timopheevii species. In T. timopheevii, BTR1-A-hapT7 is the closest wild haplotype to the domesticated haplotype BTR1-A-hap T8, differing only in the 1 bp frameshift deletion at position 567. The “Status” column refers to the historical domestication status according to the relevant seed source. Colors help to detect the differences.