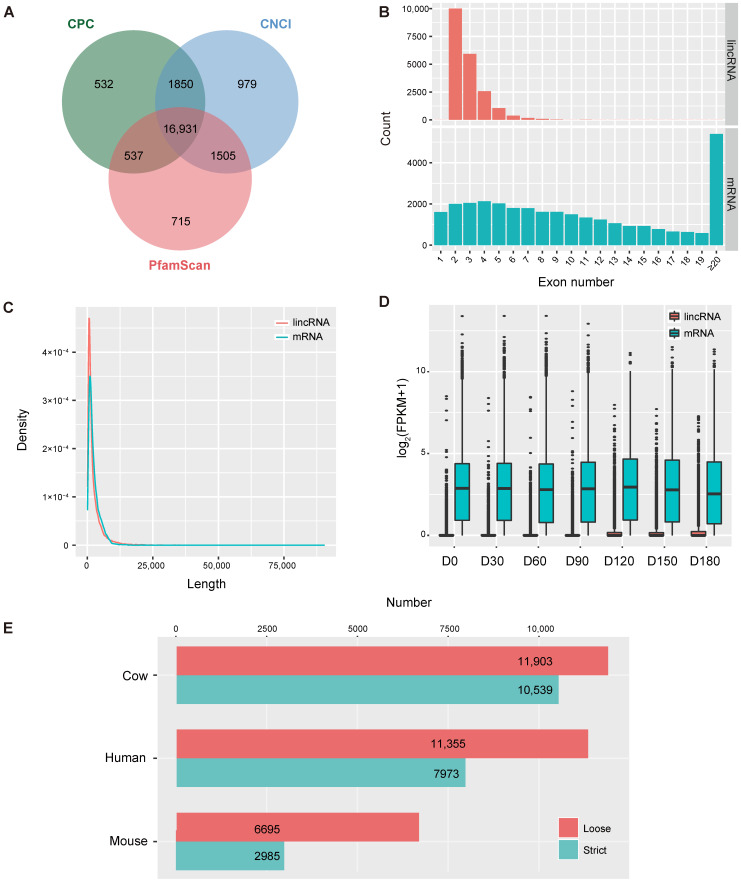
Figure 1.
Identification of long intergenic non-coding RNAs (lincRNAs). (A) Identification of the coding potential of transcripts using CNCI, CPC and PfamScan. (B) Compared with mRNAs, lincRNAs had fewer exons. (C) Compared with the mRNAs, the lincRNAs were shorter. (D) Transcriptional levels of lincRNAs and mRNAs. The transcription levels of lincRNAs and mRNAs at each age are represented by the mean value of the three individuals. (E) Number of goat testicular lincRNAs that are homologous to NONCODE lincRNAs in the cow, human, and mouse with a loose (E-value < 1 × 10−3) and strict (E-value < 1 × 10−10) threshold by BLASTN.