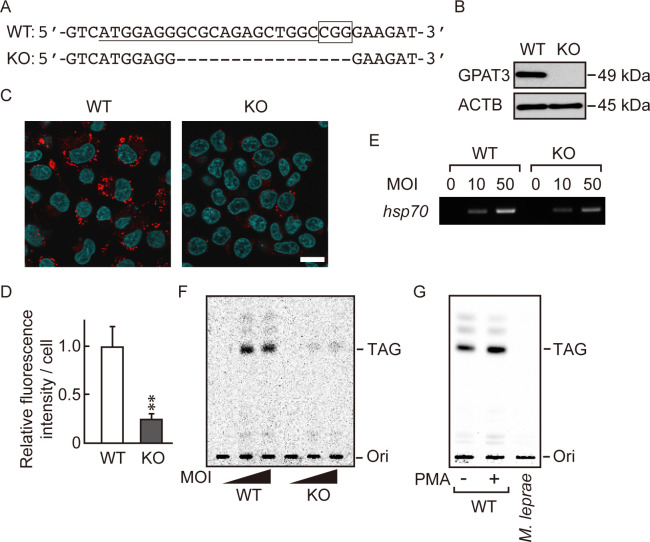
Fig 3. M. leprae utilizes host TAG synthesized via a GPAT3-dependent pathway.
(A) The 20-bp target sequence of gRNA used for the CRISPR/Cas9 gene editing system (underlined) and the PAM sequence (boxed) in wild-type (WT) GPAT3. A dashed line in the GPAT3 KO sequence indicates the frameshifting deletion as detected by DNA sequencing. (B) Western blot confirmed the absence of GPAT3 protein in KO cells. (C) LipidTOX staining (red) of WT and GPAT3 KO THP-1 cells infected or uninfected with M. leprae (MOI: 50) for 24 h with Hoechst 33258 counterstaining (blue). Scale bar: 5 μm. (D) Quantification of LipidTOX staining using fluorescence intensity values to quantify lipid droplets. Significance was determined with a Student’s t test. Two asterisks indicate p<0.01. (E and F) WT and GPAT3 KO THP-1 cells were cultured in medium containing 0.2 μCi of [14C] stearic acid for 16 h after M. leprae infection (MOI: 10 and 50). (E) M. leprae isolated from infected THP-1 cells was confirmed by PCR amplification of the M. leprae hsp70 DNA. (F) M. leprae was purified from cells, and the bacilli lipids were extracted and separated by TLC. (G) THP-1 cells were treated with PMA (20 ng/mL) for 24 h to promote lipid droplet formation, then incubated with 0.2 μCi of [14C] stearic acid for 16 h. Cell lysate was sonicated and mixed with M. leprae then incubated for 24 h. M. leprae was isolated and extracted lipids were separated by TLC to evaluate radioactivity.