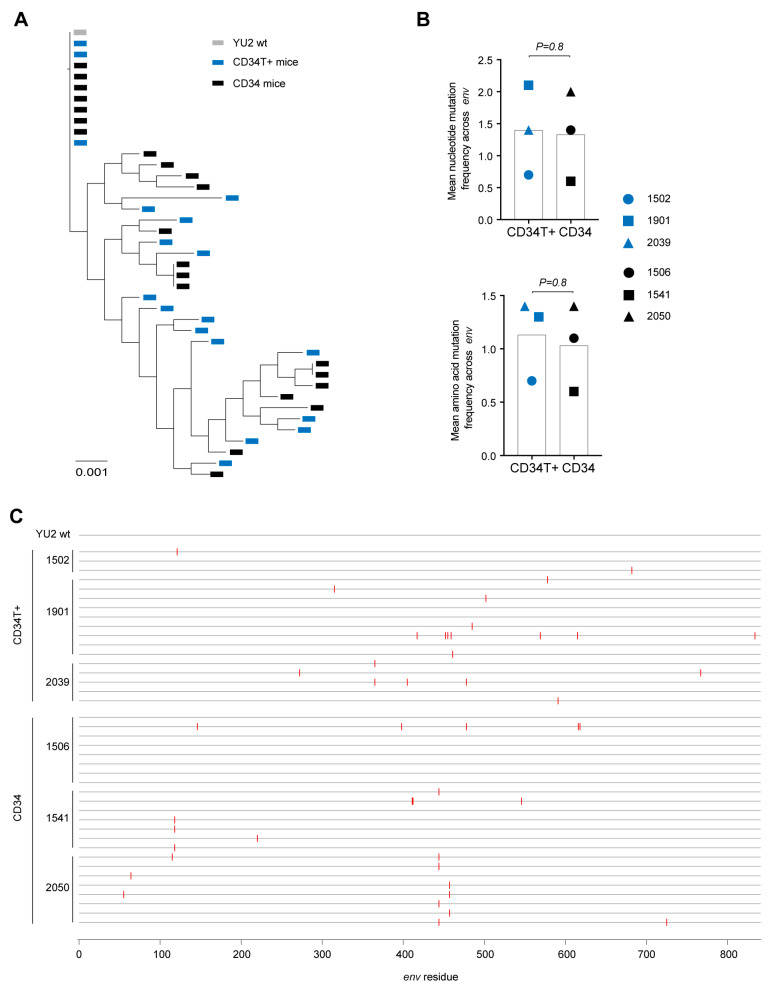
Figure 3.
In vivo diversification and evolution of HIV-1 infected mice. (A) Phylogenetic tree of SGS-derived env sequences from the plasma of CD34T+ mice and CD34 mice at day 21 post post i.r. or i.p. challenge with NL4-3YU2 HIV-1 respectively. (B) Analysis of mutation rates at the amino acid and nucleotide level in the env gene of mice from (A). (C) Amino acid alignment of plasma SGS-derived env sequences from mice analyzed in (A,B). Red bars indicate amino acid residues with mutations relative to the wild-type NL4-3YU2 HIV-1 env. Amino acid numbering is based on HIV-1YU2 env. i.r. intra-rectal, i.p. intra-peritoneal.