Figure 4. Activation of EEC Trpa1 signaling promotes intestinal motility.
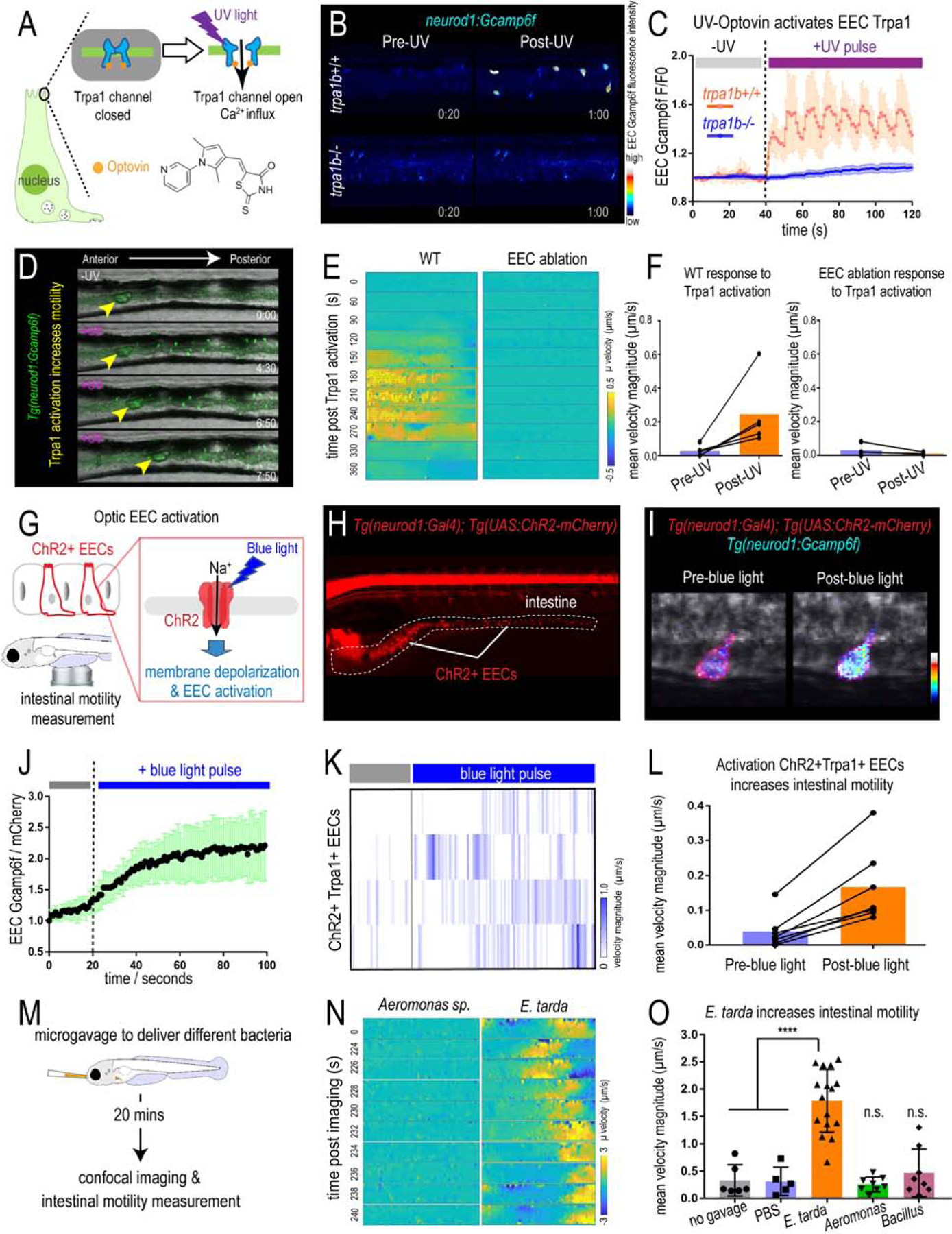
(A) Illustration of EEC Trpa1 activation using an Optovin-UV platform. (B) Confocal image of trpa1b+/+ and trpa1b−/− Tg(neurod1:Gcamp6f) zebrafish EECs before and after UV activation. (C) Quantification of EEC Gcamp6f fluorescence changes in trpa1b+/+ and trpa1b−/− zebrafish before and after UV induction. (D) Representative images of Tg(neurod1:Gcamp6f) zebrafish intestine before and after UV-induced Trpa1 activation. Yellow arrowheads indicate the movement of intestinal luminal contents from anterior to posterior following EEC activation. (E) PIV-Lab velocity analysis to quantify intestinal motility in WT and EEC ablated zebrafish. Spatiotemporal heatmap series representing the µ velocity of the imaged intestinal segment at the indicated timepoint post Trpa1 activation. (F) Quantification of the mean intestinal velocity magnitude before and after UV activation in WT and EEC ablated zebrafish. (G) Model of light activation of ChR2 in EECs. (H) Fluorescence image of Tg(neurod1:Gal4); Tg(UAS:ChR2-mCherry) zebrafish that express ChR2 in EECs. (I) Confocal image of ChR2 expressing EECs in Tg(neurod1:Gcamp6f) intestine before and after blue light-induced ChR2 activation. (J) Quantification of EEC Gcamp fluorescence intensity before and after blue light-induced ChR2 activation. (K) Intestinal velocity magnitude before and after blue-light induced activation in ChR2+Trpa1+ EECs. (L) Mean velocity magnitude before and after blue light-induced activation in ChR2+Trpa1+ EECs. (M) Experimental design schematic for panels N and O. (N) Heatmap representing the µ velocity of the imaged intestinal segment at indicated timepoints following Aeromonas sp. or E. tarda gavage. (O) Mean intestinal velocity magnitude in zebrafish without gavage or gavaged with PBS or different bacterial strains. Student’s t-test was used in O. ****p<0.0001.
