Fig. 2. Selective decrease in tRNAPhe in Ftsj1 KO mouse brain.
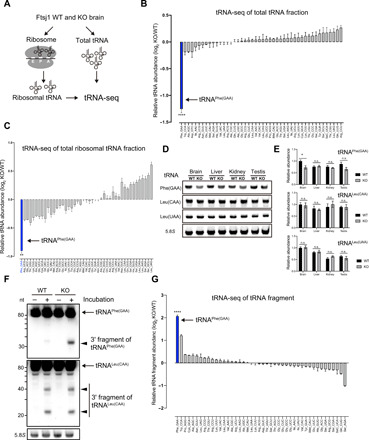
(A) Schematic illustration of tRNA-seq strategy for analysis of total tRNA and ribosome-bound tRNA isolated from WT and KO mouse brains at 8 weeks. (B) Relative abundance of individual tRNA in the KO mouse brain. tRNAPhe of the KO mouse brain showed the most pronounced decrease compared to that of WT mouse brain (n = 3 for each, ****P < 0.0001 by Student’s t test). (C) Relative abundance of individual ribosome-bound tRNA. Among all tRNAs in the KO mouse brain, tRNAPhe showed the largest reduction (n = 3 for each, **P = 0.0019 by Student’s t test). (D) Northern blotting of tRNAPhe, tRNALeu(CAA), and tRNALeu(UAA) from total RNA isolated from the brain, liver, kidney, and testis of 8-week-old WT and KO mice. 5.8S ribosomal RNA (rRNA) was used as loading control. (E) Quantitative analysis of tRNAPhe, tRNALeu(CAA), and tRNALeu(UAA) level in indicated tissues relative to the level in the WT brain. tRNAPhe was selectively and significantly decreased in the brain, but not in the liver, kidney, and testis (n = 4 for each, *P = 0.02 by Student’s t test). n.s., not significant. (F) Lysates of WT and KO mouse brains were briefly incubated on ice and subjected to RNA extraction and Northern blotting against tRNAPhe and tRNALeu(CAA). 5.8S rRNA was used as loading control. Note that a 3′ fragment of tRNAPhe was strongly detected in KO mouse brain lysate. (G) Relative abundance of tRNA fragments in WT and KO mouse brains were analyzed by tRNA-seq. Note that the tRNAPhe-derived fragment was the most abundant tRNA species that accumulated in the KO mouse brain when compared to WT (n = 3 for each, ****P < 0.0001 by Student’s t test). Error bars present SEM.
