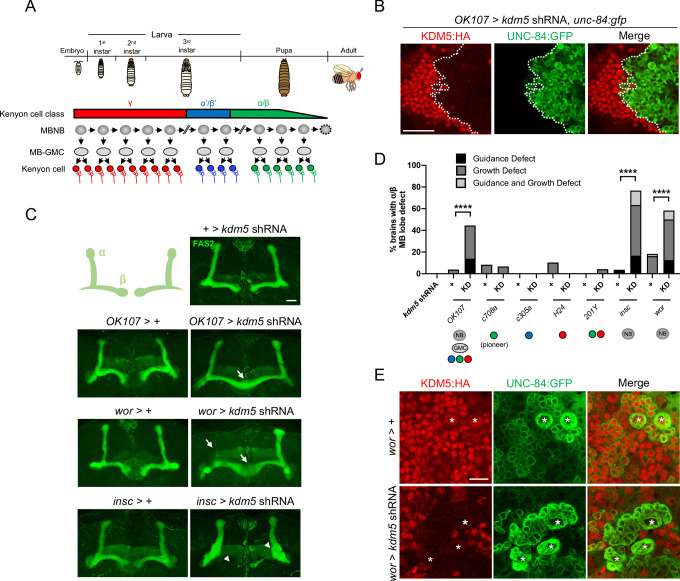
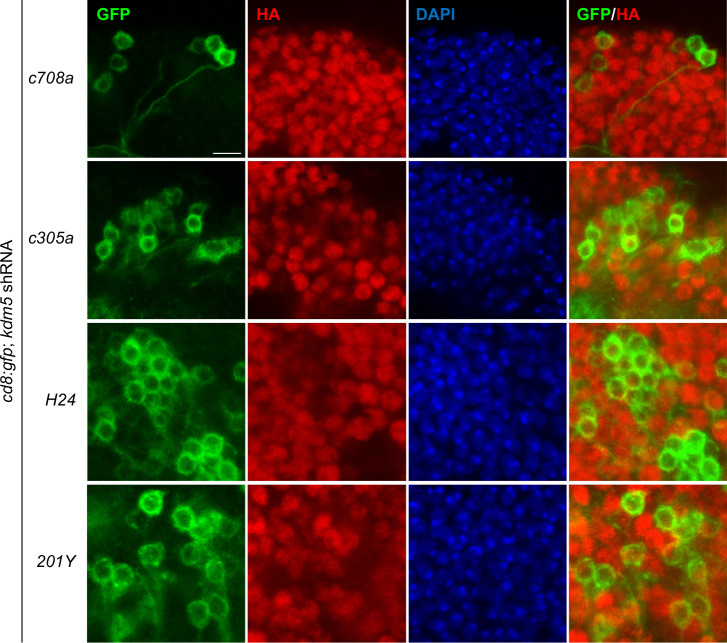
Figure 3. Depletion of lysine demethylase 5 (KDM5) within neural precursors results in neuroanatomical defects of the mushroom body (MB).
(A) Schematic showing the sequential generation of distinct subclasses of Kenyon cells throughout development. MB neuroblasts (MBNBs) are eliminated via apoptosis (dotted line) immediately prior to eclosion. (B) OK107-Gal4 driving expression of UNC-84:GFP to reveal shRNA-mediated KDM5:HA depletion within adult Kenyon cell nuclei. (C) Representative Z projections of adult kdm5 knockdown animals exhibiting significant α/β lobe defects and their respective kdm5 shRNA and GAL4 controls. The antibody anti-fasciclin 2 is used to visualize α/β lobes. Arrows indicate growth defects, and arrowheads indicate guidance defects. (D) Quantification of α/β MB lobe defects in flies expressing kdm5 shRNA driven by neural progenitor cell- and Kenyon cell-specific drivers. ‘KD’ indicates shRNA-mediated knockdown of kdm5. n = 16–49 (mean n = 29). ****p<0.0001 (chi-square test with Bonferroni correction). (E) Z projection of larval cortex revealing wor-Gal4-driven expression of kdm5 shRNA and unc-84:gfp transgenes. KDM5:HA depletion is observed in presumptive ganglion mother cells and post-mitotic cells surrounding NBs (marked by asterisks). Scale bars represent 20 μm in (B) and (C) and 10 μm in (E).