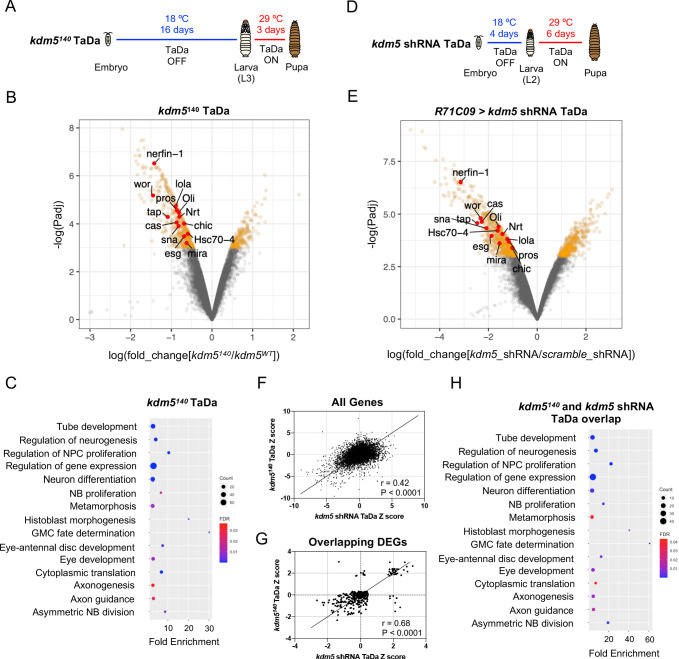
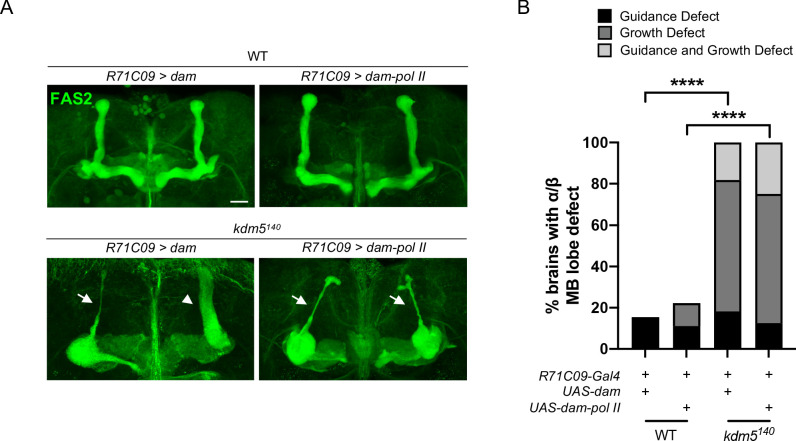
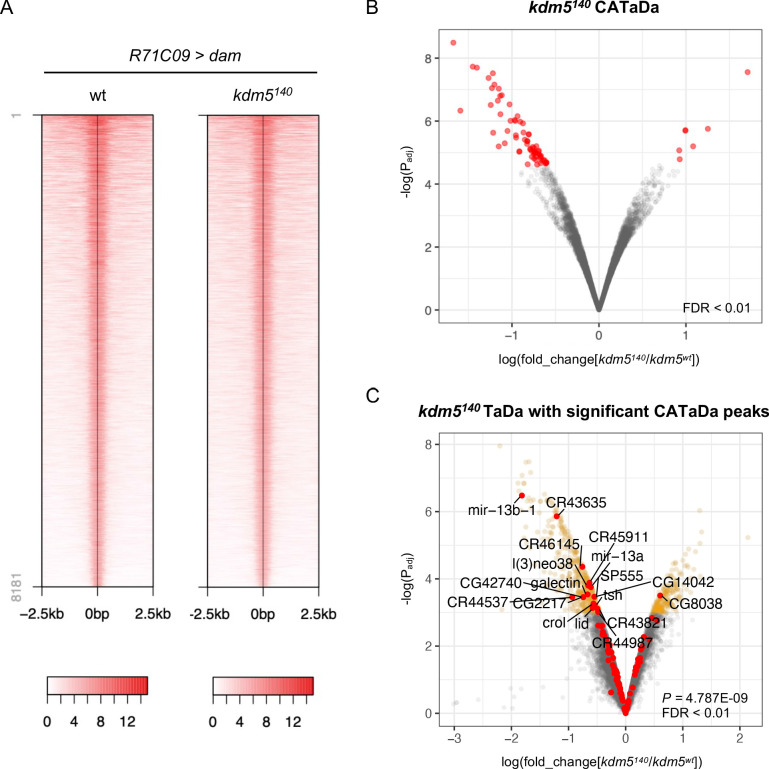
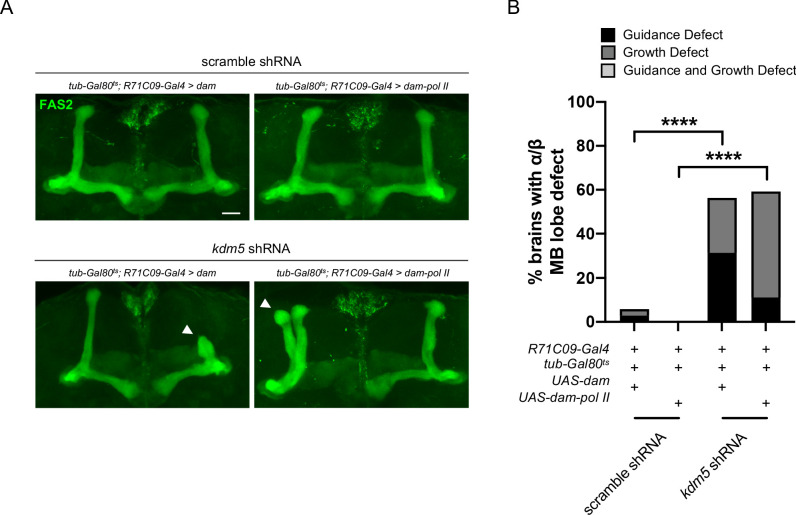
Figure 5. Transcriptome profiling of lysine demethylase 5 (KDM5)-depleted ganglion mother cells (GMCs) and immature neurons by targeted DamID (TaDa) reveals KDM5-regulatory networks critical for ganglion mother cell (GMC) proliferation and neurodevelopment.
(A) Timeline of TaDa induction within GMCs and immature neurons of kdm5140 WL3 animals and pupae. (B) Volcano plot of differentially expressed genes (DEGs) within GMCs and immature neurons of kdm5140 animals compared to wild type. Genes with a false discovery rate (FDR) < 0.05 are in red, with those labeled involved in GMC proliferation and neurodevelopment. TaDa analyses were performed in quintuplicate. (C) Representative distribution of ontology terms for DEGs in kdm5140 GMCs and immature neurons using a PANTHER Overrepresentation Test (Fisher’s exact test with FDR < 0.05). (D) Timeline of TaDa and kdm5 shRNA induction within GMCs and immature neurons of WL3 animals and pharate adults. (E) Volcano plot of DEGs within kdm5 shRNA GMCs and immature neurons compared to those expressing a scrambled shRNA. Genes with an FDR < 0.05 are in red, with those labeled involved in GMC proliferation and neurodevelopment. TaDa analyses were performed in triplicate. (F) Correlation of Z scores between DEGs of kdm5140 and kdm5 shRNA TaDa datasets (Deming regression; p<0.0001). (G) Correlation of Z scores between overlapping DEGs of kdm5140 and kdm5 shRNA TaDa datasets (Deming regression; p<0.0001). (H) Representative distribution of ontology terms for DEGs in overlapping kdm5140 and kdm5 shRNA TaDa datasets utilizing a PANTHER Overrepresentation Test (Fisher’s exact test with FDR < 0.05).