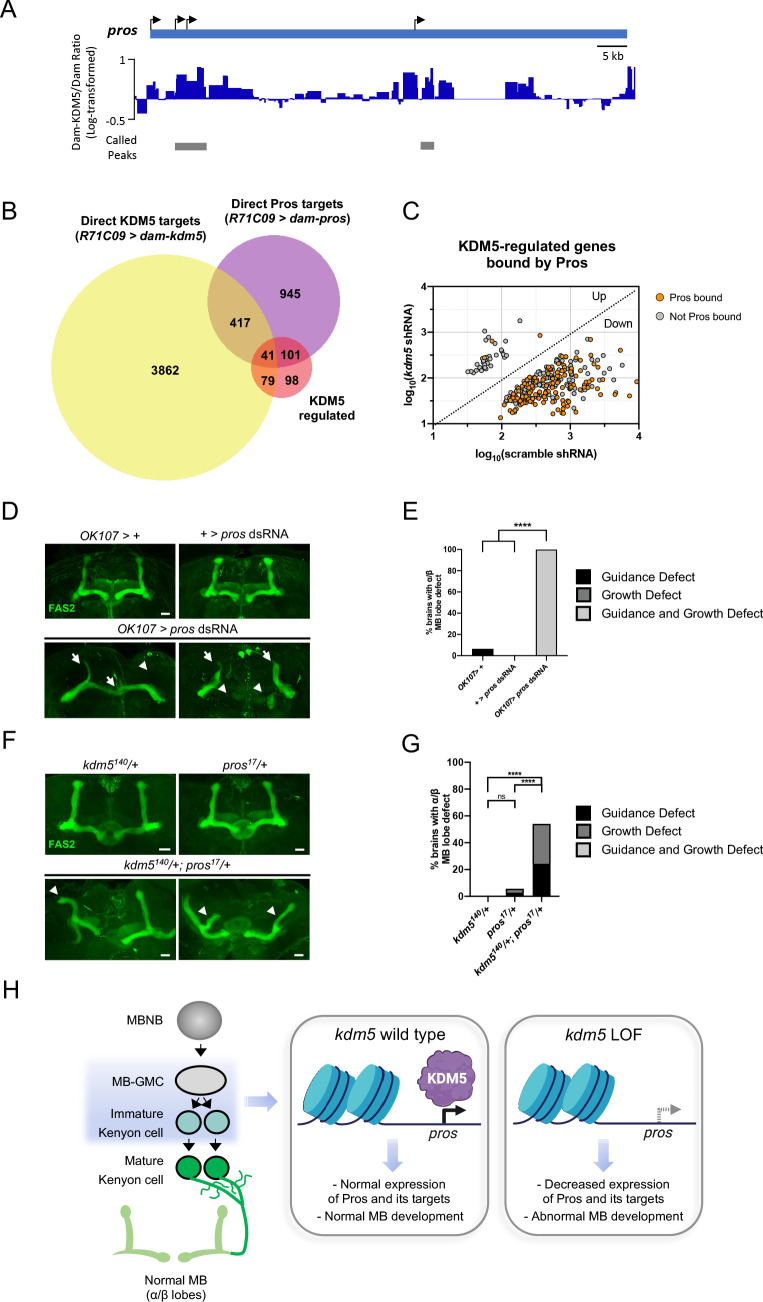
Figure 7. Neuromorphological and transcriptomic analyses reveal a genetic interaction between prospero and lysine demethylase 5 (KDM5).
(A) Integrative genomics viewer (IGV) plot showing average KDM5 occupancy at pros transcriptional start sites across six replicates. Scale bars represent the log2 ratio change between Dam-KDM5 and Dam samples. Called peaks are indicated by gray bars. (B) Venn diagram illustrating intersection of similarly dysregulated kdm5140 and kdm5 shRNA overlapping differentially expressed genes (DEGs) with Dam-KDM5 direct binding data and a previously published Dam-Pros targeted DamID (TaDa) binding dataset (Fisher’s exact test, p=2.20E-16 for KDM5-dysregulated and Pros gene overlap) (Liu et al., 2020). (C) Analysis of the 319 similarly dysregulated kdm5140 and kdm5 shRNA overlapping DEGs (with values plotted from the kdm5 shRNA TaDa). Direct Pros targets from the previously published Dam-Pros TaDa dataset (Liu et al., 2020) are labeled in orange. (D) Representative Z projections of OK107 > pros RNAi adults exhibiting significant α/β lobe defects. The α/β lobes are revealed by anti-fasciclin 2 (Fas2). (E) Quantification of α/β mushroom body (MB) lobe defects in flies expressing pros shRNA driven by OK107-Gal4. n = 16–19 (mean n = 17). ****p<0.0001 (chi-square test with Bonferroni correction). (F) Representative Z projections of representative kdm5140/+ and pros17/+ heterozygous adult α/β MB lobes (top) and kdm5140/+; pros17/+ transheterozygous adult α/β MB lobes (bottom). The α/β lobes are revealed by anti-Fas2. (G) Quantification of α/β MB lobe defects in kdm5140/+; pros17/+ transheterozygous adults and heterozygous controls. n = 28–37 (mean n = 34). ****p<0.0001 (chi-square test with Bonferroni correction). (H) Model proposing a genetic interaction between pros and kdm5 within ganglion mother cells (GMCs) and immature α/β Kenyon cells. KDM5 binds to the pros locus and positively regulates its transcription. Loss of kdm5 leads to downregulation of pros and its targets, resulting in defects to MB neurodevelopment and cognitive function. Image was created with BioRender and Venn diagrams with BioVenn (Hulsen et al., 2008). Scale bars represent 20 μm.