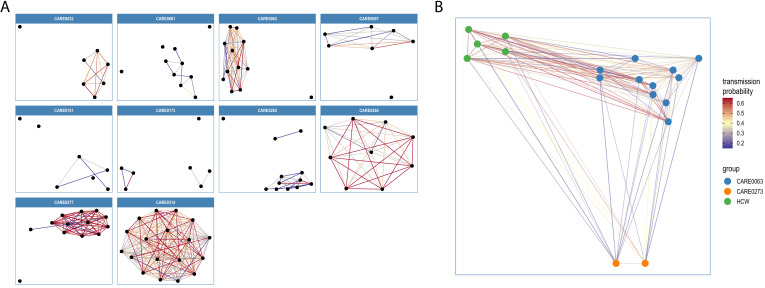
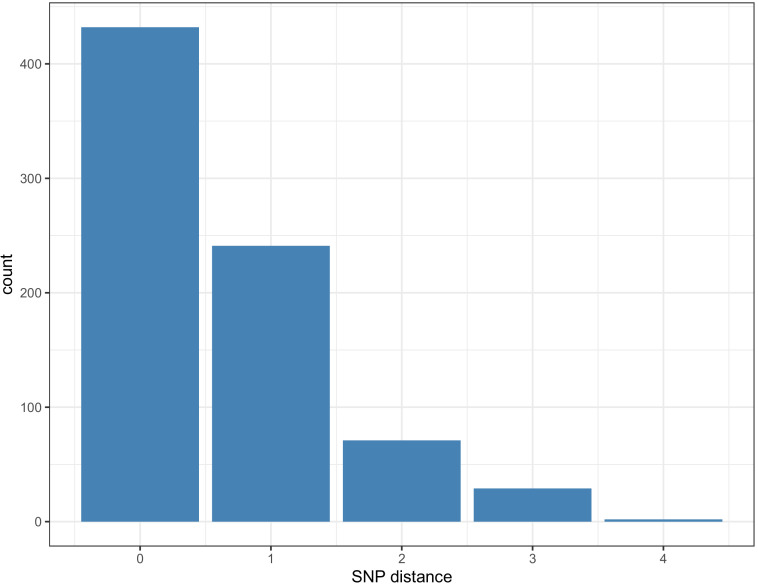
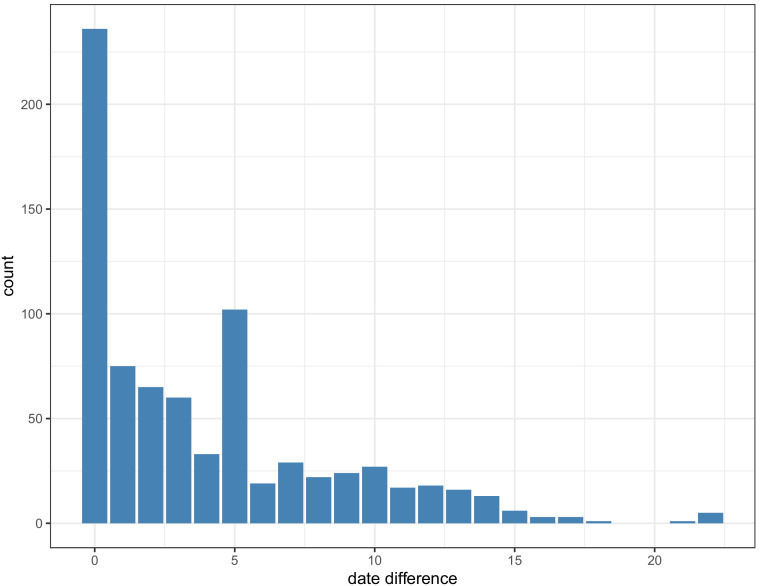
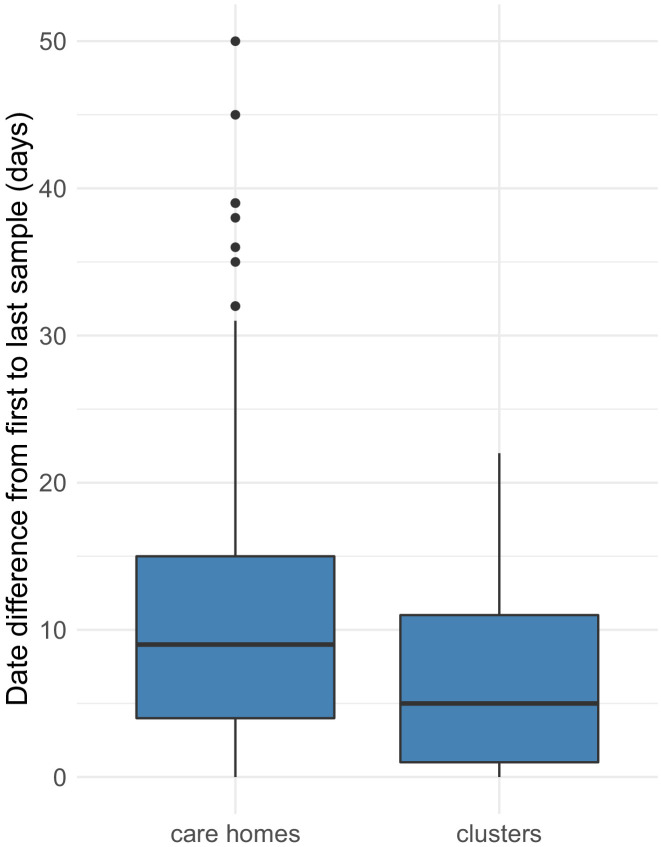
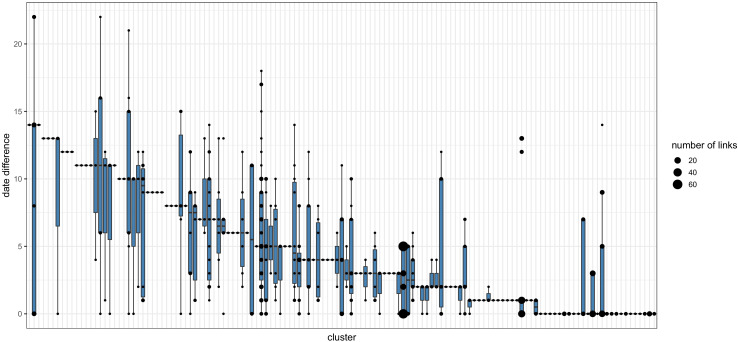
Figure 7. Visualisations of SARS-CoV-2 clusters among care home residents.
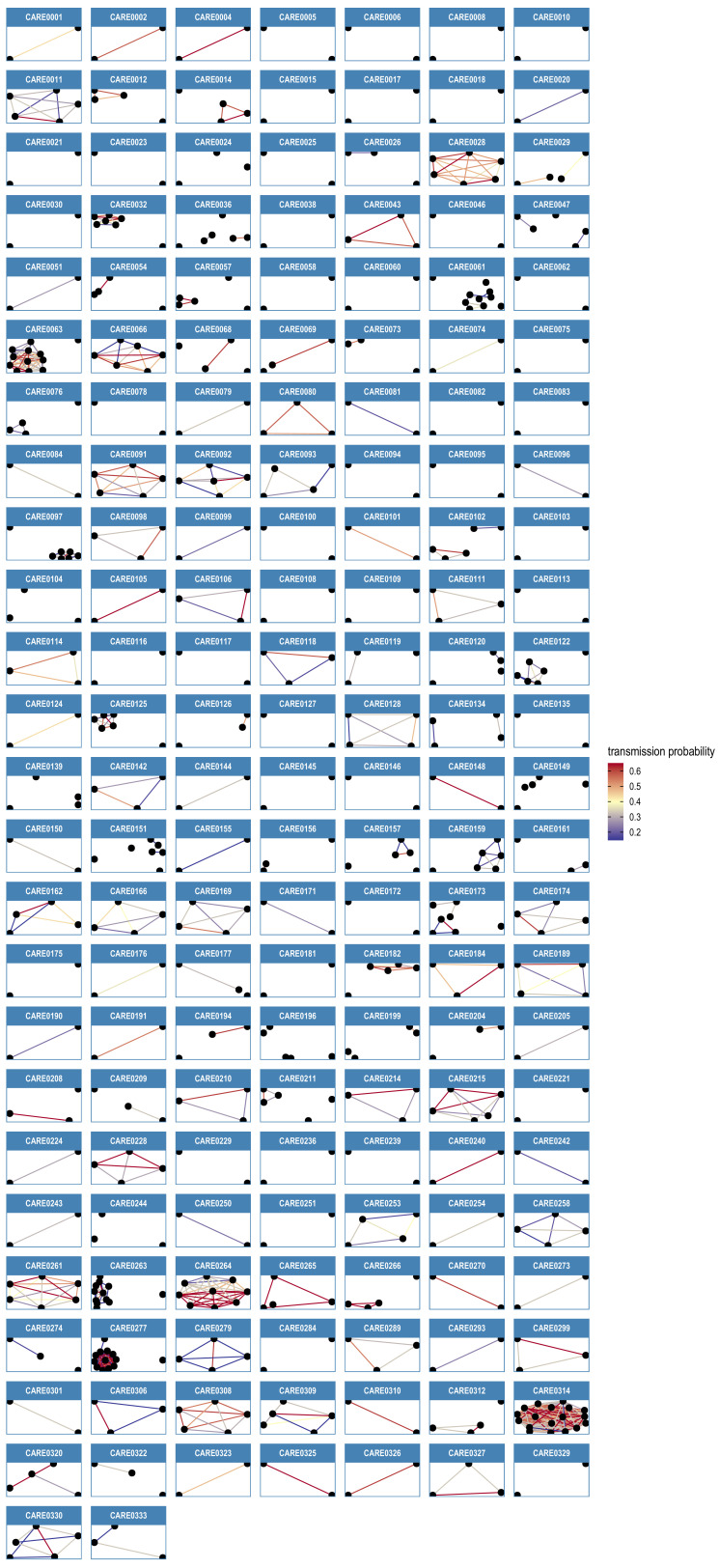
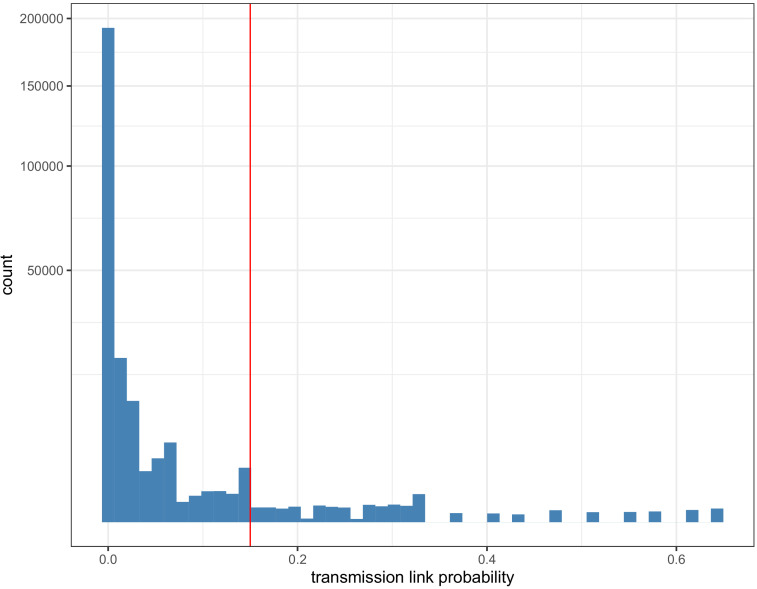
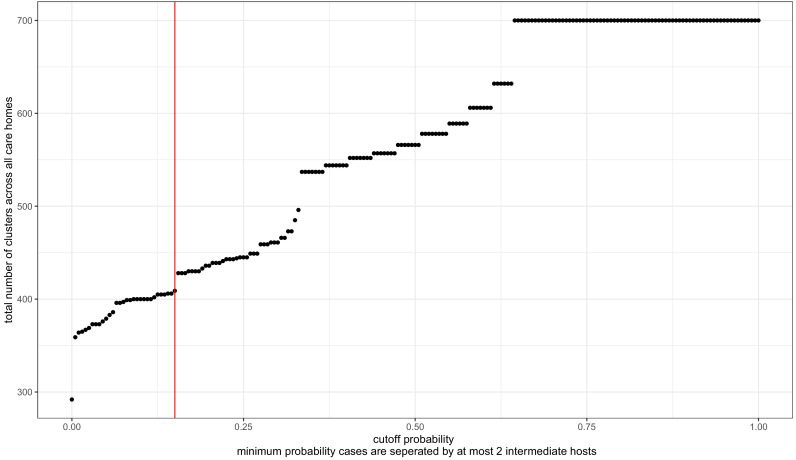
Transmission networks were produced using a derivative of the transcluster algorithm, which incorporates pairwise date and genetic differences to estimate the probability of cases being connected within a defined number of intermediate hosts. Clusters were defined using a probability threshold of >15% for cases being linked by <2 intermediate hosts (further details in Materials and methods). (A) Transmission clusters for the ten care homes with the largest number of care home residents with available genomes. Consistent with Figure 6, several of the 10 care homes with the largest number of genomes comprised single transmission clusters (e.g. CARE0314), while others contained two or more clusters consistent with multiple independent transmission sources among the residents. These data alone do not indicate where the residents acquired their infections, and hospital-acquired infections for some of the clusters is a possibility alongside multiple introductions into the same care homes. (B) Visualisation of transmission links between residents of two nearby carehomes and a group of healthcare workers (HCW). Two care homes, CARE0063 (blue) and CARE0273 (orange), each had strong transmission links identified with the transcluster algorithm to a group of HCW (green). The HCW comprised paramedics and care home carers – one working at CARE0063 and the other working at an unknown care home. We do not have confirmatory epidemiological data available, but this raises the possibility of the cases sharing a linked transmission network.