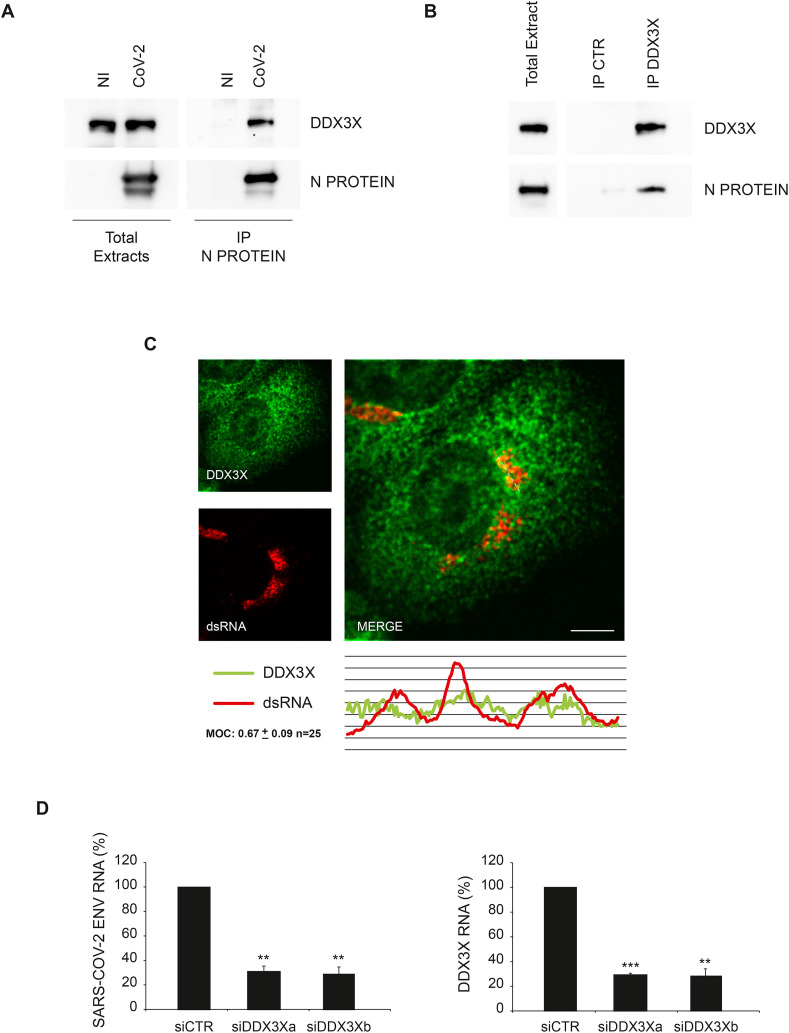
Fig. 3.
SARS-CoV-2 Nucleoprotein interacts with DDX3X. A) Vero E6 cells infected with SARS-CoV-2 at MOI 0.01 were harvested after 24 h and protein extracts were subjected to immunoprecipitation using an anti-SARS-CoV Nucleoprotein antibody. Immunopurified complexes were analyzed by immunoblotting using anti-SARS-CoV Nucleoprotein and anti-DDX3X antibodies. B) Vero E6 cells infected with SARS-CoV-2 at MOI 0.01 were harvested after 24 h and protein extracts were subjected to immunoprecipitation using an anti-DDX3X antibody or an anti-IgG antibody as a negative control (IP CTR). Immunopurified complexes were analyzed by immunoblotting using anti-SARS-CoV Nucleoprotein and anti-DDX3X antibodies. C) Vero E6 cells infected with SARS-CoV-2 at MOI 0.01 were fixed after 24 h and analyzed for DDX3X and dsRNA localization by immunofluorescence using specific antibodies. Bottom panel shows colocalization trace profiles. Colocalization rate was measured by Mander's overlap coefficient (MOC) on 25 cells by using ImageJ software. NI: non-infected; N PROTEIN: Nucleoprotein; CoV-2: SARS-CoV-2; CTR: unrelated IgG; dsRNA: double strand RNA. Scale bar, 5 μm. D) Vero E6 cells were transfected with small interfering RNAs specific for DDX3X (siDDX3Xa and siDDX3Xb), or non-targeting siRNA (siCTR) as negative control, and infected with SARS-CoV-2 at MOI 0.001. Twenty-four hours later, cells were harvested and DDX3X and viral RNAs were analyzed by RT-qPCR. The graphs report means ± SD of normalized values from three independent experiments. ** = p < 0.01, *** = p < 0.001. Viral RNA levels are reported as fold changes with respect to the amount detected at 24h post infection.