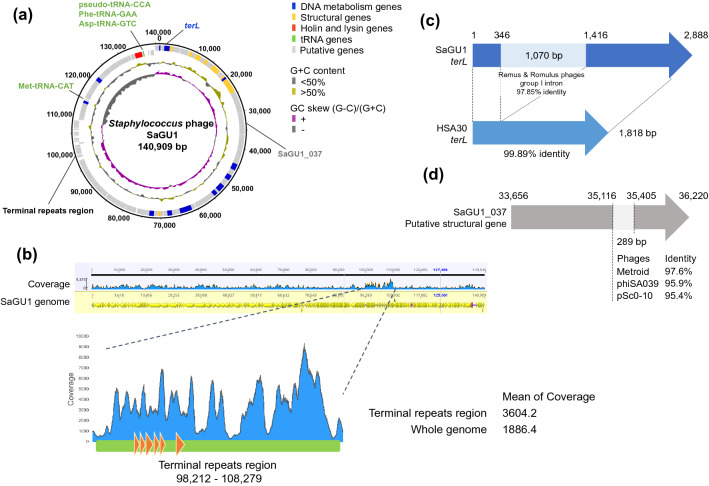
Fig. 1.
Genomic structure of the phage SaGU1. a Circular representation of the SaGU1 genome. Concentric rings denote the following features (from the outer to inner rings): nucleotide positions are forward strand (outer) and reverse strand (inner); the predicted genes are DNA metabolism genes (blue), structural genes (yellow), holin and lysin genes (red), tRNA (green), and putative genes (light gray); G + C content is < 50% (gray), > 50% (gold); GC skew is (G − C)/(G + C) (gray,−; purple, +). b Reads mapped onto SaGU1 genome sequence. Predicted terminal repeats region (green) and terminal-repeat encoded proteins (orange) c Structure of SaGU1 terL. Pale blue line represents the region with high sequence similarity to the group I intron of Staphylococcus phages Remus and Romulus. d Structure of the SaGU1_037 putative structural gene. Pale gray line represents the region with high sequence similarity to sequences of Staphylococcus phages Metroid, phiSA039, and pSc0-10