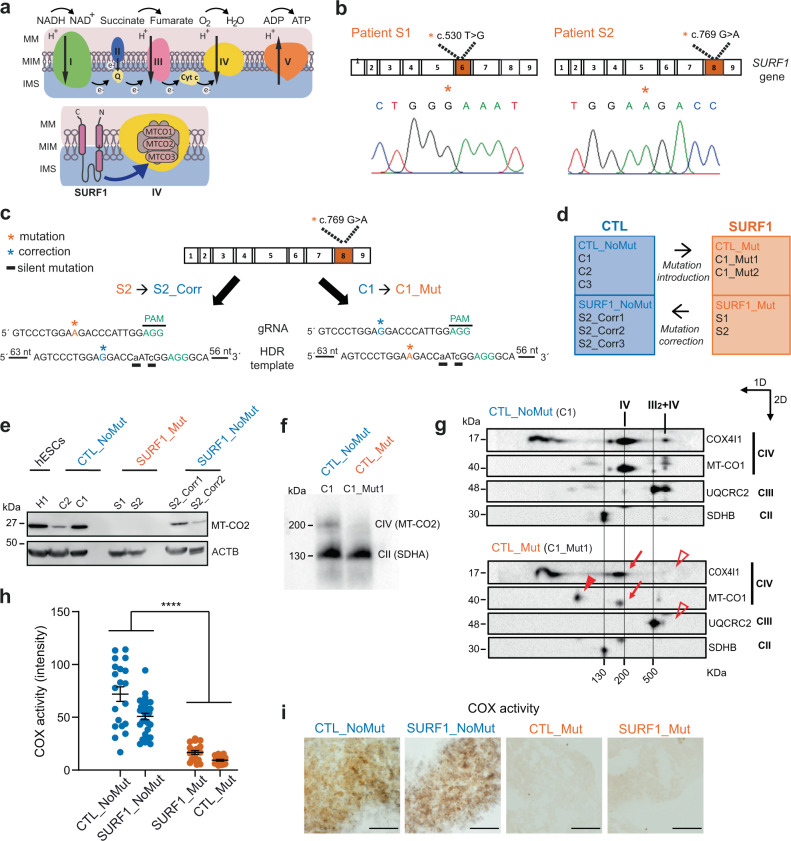
Fig. 1. Generation of an iPSC-based model of LS due to SURF1 mutations.
a Mitochondrial respiratory chain and putative SURF1 function in CIV assembly. MM mitochondrial membrane, IMM inner mitochondrial membrane, IMS intermembrane space. b Electropherograms showing 5′ > 3′ sequences of SURF1 in iPSCs from patients S1 and S2 carrying the mutations c.530T > G p.(V177G) and c.769G > A p.(G257R), respectively. Orange stars: mutation site. c Genome editing approach to correct mutation c.769G > A in patient line S2, and to introduce the same mutation in control line C1. Orange stars: mutation site; blue stars: correction site; black underlines: artificially introduced silent mutations; gRNA: guide RNA; HDR: homology direct repair. d Overview of iPSC lines used in this study. e Immunoblot of MT-CO2 (26 kDa) in NPCs derived from hESCs (H1), CTL (CTL_NoMut: C2, C1; SURF1_NoMut: S2_Corr1, S2_Corr2), and SURF1 (SURF1_Mut: S1, S2) (n = 2 independent experiments). f One dimensional (1D) blue-native gel electrophoresis (BNGE) analysis of mitochondrial complexes showing lack of incorporation of MT-CO2 into CIV in NPCs from CTL (CTL_NoMut: C1) and SURF1 (CTL_Mut: C1_Mut1). The structure of the complex II (CII) (visualized by SDHA antibody) was not affected (n = 2 independent experiments). g Two-dimensional (2D) BNGE showing mitochondrial complex assembly in NPCs from CTL (CTL_NoMut: C1) and SURF1 (CTL_Mut: C1_Mut1). Red arrows: structurally impaired CIV migrating at lower molecular weight (detected by MT-CO1 and COX4I1 antibodies); red arrowhead: accumulation of COX assembly intermediates (detected by MT-CO1 antibody); open red arrowheads: loss of III2 + IV supercomplex (detected by COX4I1 and UQCRC2 antibodies). We used CIII (detected by UQCRC2 antibody) and CII (detected by SDHB antibody) to align CTL blot with SURF1 blot (n = 2 independent experiments). h, i Quantification and representative images of COX activity intensity in CTL NPCs (CTL_NoMut: C1, C2, C3; SURF1_NoMut: S2_Corr1, S2_Corr2) and SURF1 NPCs (SURF1_Mut: S1, S2; CTL_Mut: C1_Mut1) (mean ± s.e.m.; each dot represents a biological replicate; n = 10 biological replicates per line over three independent experiments; ****p < 0.0001 CTL vs. SURF1; two-sided Mann–Whitney U test). Scale bar: 500 nm.