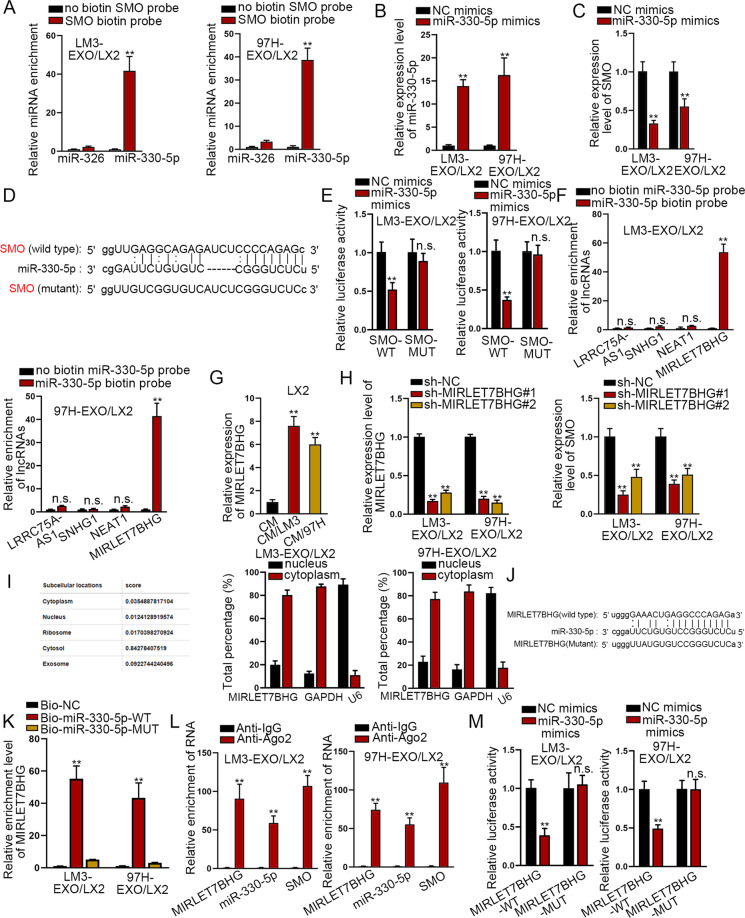
Fig. 4. MIRLET7BHG sponged miR-330-5p to upregulate SMO.
A RNA pull-down assay examined the enrichment of miR-326 and miR-330-5p by biotin-labeled SMO. B qRT-PCR examined the overexpression efficiency of miR-330-5p in activated HSCs. C qRT-PCR revealed the influence of upregulated miR-330-5p on SMO expression in activated HSCs. D Binding sites between SMO and miR-330-5p were predicted by starBase database. E Luciferase reporter assay examined the changes in luciferase activity of wild-type and mutant SMO in activated HSCs after overexpressing miR-330-5p. F RNA pull-down assay revealed the enrichment of four potential lncRNAs by biotin-labeled miR-330-5p. G qRT-PCR determined MIRLET7BHG expression in quiescent and activated HSCs. H qRT-PCR validated the depletion efficiency of MIRLET7BHG and also evaluated the influence of silenced MIRLET7BHG on SMO expression in activated HSCs. I LncLocator database and subcellular fraction assay revealed the subcellular location of MIRLET7BHG in activated HSCs. J Binding sites between MIRLET7BHG and miR-330-5p were predicted by starBase database. K RNA pull-down assay revealed the enrichment of MIRLET7BHG pulled down by biotin-labeled miR-330-5p. L RIP assay analyzed the enrichment of MIRLET7BHG, miR-330-5p, and SMO in anti-Ago2 group relative to anti-IgG group. M Luciferase reporter assay detected the changes in luciferase activity of wild-type and mutant MIRLET7BHG under miR-330-5p overexpression. **P < 0.01, “n.s” represents no significance.