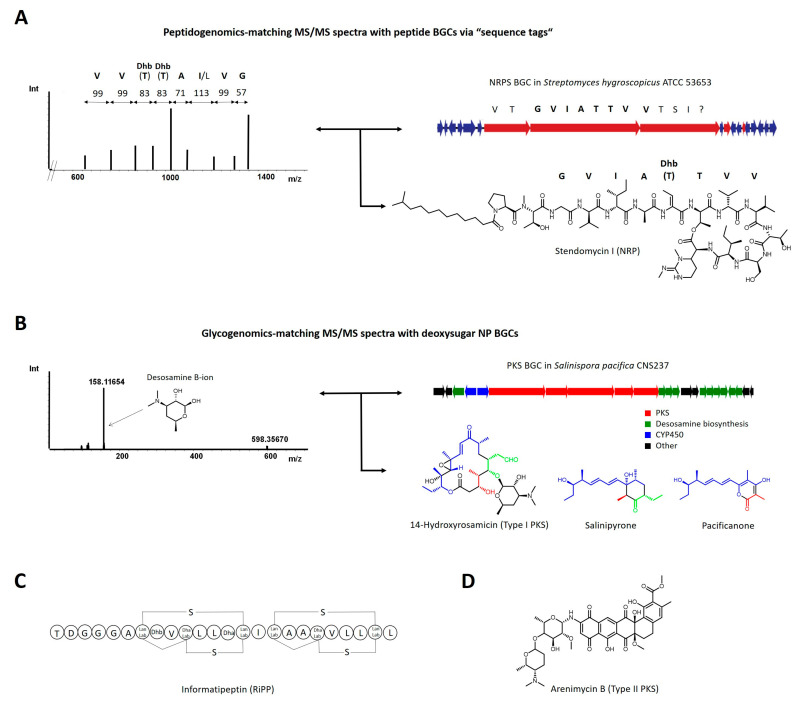
Figure 1.
Concepts and discovery examples for experiment-guided genome mining. (A) Peptidogenomics: Streptomyces hygroscopicus MS/MS data yielded a sequence tag with eight amino acids, two of them dehydrated threonines (Dhb). The sequence tag matched with a sequence of adenylation domain predictions of an orphan nonribosomal peptide synthetase BGC, facilitating the targeted isolation and characterization of stendomycin lipopeptides [22]. The second threonine dehydration appeared only during MS/MS fragmentation as elimination product of the ester bond. (B) Glycogenomics: Matching of MS/MS spectra of Salinispora pacifica CNS237 with a type I polyketide synthase BGC encoding deoxysugar biosynthesis genes revealed several rosamicin derivatives and enabled their targeted isolation and further characterization. The previously isolated linear polyketides salinipyrone and pacificanone appear to be shunt products of the rosamicin PKS, revealed by mutagenesis experiments. Building blocks synthesized by the same module(s) are color-coded accordingly [33]. (C) Further natural products from different classes discovered by the peptidogenomic [28] and (D) glycogenomic approach [23]. For more details regarding these concepts, please refer to references [22,23].