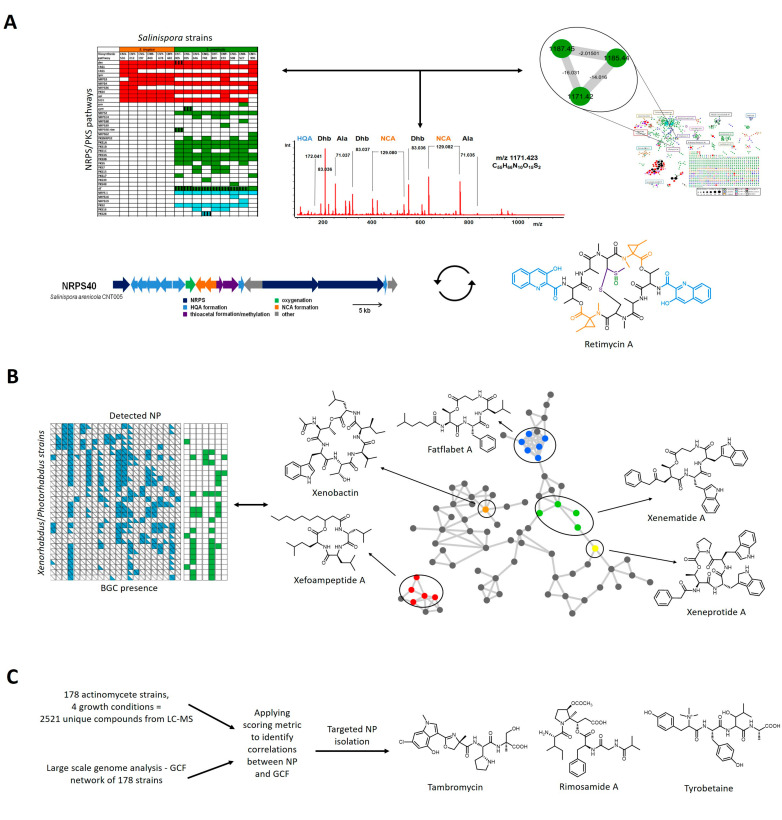
Figure 2.
Discovery examples for correlation-based approaches using paired datasets. (A) Correlation of Salinispora strain BGC patterns with a molecular network of 35 strains led to the identification of a candidate peptide, encoded and produced by only one strain in the dataset. The peptide was matched to its NRPS BGC with additional help of the peptidogenomic approach. The structure of the elucidated metabolite retimycin A is depicted. Building blocks are color-coded corresponding to responsible biosynthetic genes [39]. Taken from reference 39 and rearranged with permission of Elsevier. (B) Xenorhabdus and Photorhabdus strains were analyzed for BGC patterns and production of encoded metabolites (left). Subsequent molecular network analysis led to the identification and discovery of several NRPS-derived cyclic depsipeptides (right) [40]. (C) Metabologenomic workflow of a 178 strain actinomycetes dataset [41,42,43,44]. An example for the applied scoring metric can be found in reference 21.