Figure 1.
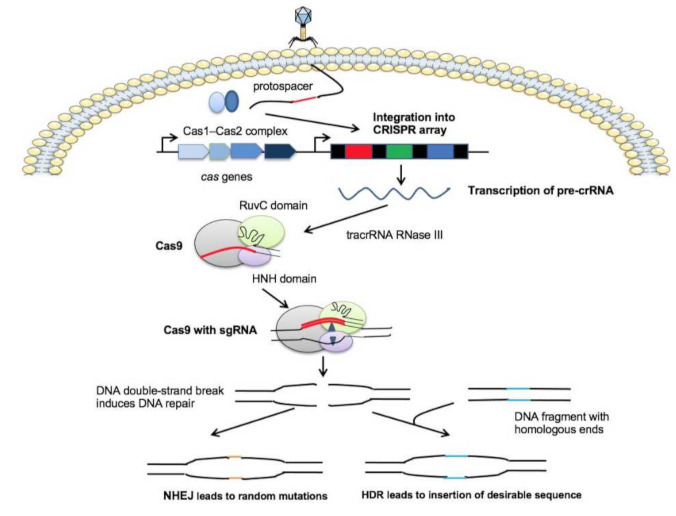
Molecular mechanism of the CRISPR-Cas 9 system. CRISPR-Cas systems are composed of a cas operon (blue arrows) and a CRISPR array which identical repeat sequences (black rectangles) that are interspersed by phage-derived spacers (colored rectangles). Upon phage infection, a sequence of the invading DNA (protospacer) is incorporated into the CRISPR array by the Cas1-Cas2 complex. The CRISPR array is then transcribed into a long precursor CRISPR RNA (pre-crRNA). In the CRISPR-Cas9 system, crRNA maturation requires tracrRNA, RNase III, and Cas9. The Cas9 protein contains two nuclease domains, the RuvC domain, and the HNH domain. Cas9 is guided by a sgRNA to induce a double-strand DNA break at a desired genomic locus. The sgRNA is composed of tracrRNA and crRNA. The tracrRNA hybridizes to the crRNA and binds to the Cas9 protein, forming the CRISPR-CAs9/sgRNA complex to edit genome sequences. DNA damage can be repaired by nonhomologous end joining (NHEJ), yielding short random insertions or deletions at the target site. Alternatively, a DNA sequence that shows partial complementarity to the target site can be inserted during homology-directed repair (HDR) for precise genome editing purposes.
