Abstract
Most species in the genus Amanita are ectomycorrhizal fungi comprising both edible and poisonous mushrooms. Some species produce potent cyclic peptide toxins, such as α-amanitin, which places them among the deadliest organisms known to mankind. These toxins and related cyclic peptides are encoded by genes of the “MSDIN” family (named after the first five amino acid residues of the precursor peptides), and it is largely unknown to what extent these genes are expressed in the basidiocarps. In the present study, Amanita rimosa and Amanita exitialis were sequenced through the PacBio and Illumina techniques. Together with our two previously sequenced genomes, Amanita subjunquillea and Amanita pallidorosea, in total, 46 previously unknown MSDIN genes were discovered. The expression of over 80% of the MSDIN genes was demonstrated in A. subjunquillea. Through a combination of genomics and mass spectrometry, 12 MSDIN genes were shown to produce novel cyclic peptides. To further confirm the results, three of the cyclic peptides were chemically synthesized. The tandem mass spectrometry (MS/MS) spectra of the natural and the synthetic peptides shared a majority of the fragment ions, demonstrating an identical structure between each peptide pair. Collectively, the results suggested that the genome-guided approach is reliable for identifying novel cyclic peptides in Amanita species and that there is a large peptide reservoir in these mushrooms.
Keywords: poisonous mushroom, genome, MSDIN family, Amanitin, LC–HRMS
1. Introduction
The genus Amanita (Persoon 1797) is double-faced: on one hand, it harbors some of the best-known gourmet mushrooms such as Amanita caesarea, once favored by emperors of Rome [1]; on the other hand, it causes over 90% of deadly mushroom poisonings worldwide because most have little knowledge to distinguish the edible from the deadly [2,3,4]. The majority of the species within this genus are important ectomycorrhizal fungi that have associations with more than ten families of trees, and they play important roles in the ecosystem health of forests [5].
Research on toxins in amanitin-producing (referred as deadly or lethal in this report) Amanita mushrooms dates back to the 19th century. Due to technical difficulties in extracting, purifying, and analyzing the chemical substances, significant advances were not made until the 20th century [6]. Lynen and Wieland purified one of the major toxins, phalloidin, in 1938 [7]. Then, α-amanitin was purified and crystalized three years later [8]. The death cap, Amanita phalloides, became well-known for its deadly poison in 1951 from death reports [9]. As research has progressed, two major types of toxins, i.e., amatoxins and phallotoxins, have been found in dozens of lethal Amanita species. The amatoxins function as a highly specific, efficient inhibitors of eukaryotic RNA polymerase II [10,11], and the phallotoxins effectively prevent the depolymerization of actin fibers [12,13,14]. It has been shown that amatoxins and phallotoxins in Amanita are synthesized on ribosomes, which represents the first ribosomal cyclic peptide discovered in the Fungi kingdom [15]. They are encoded by the “MSDIN” gene family as precursor peptides of 34–37 amino acids. The precursors are then cleaved and macrocyclized into 7–10 amino acid cyclic peptides by a specialized prolyl oligopeptidase B (POPB) [16,17]. It is a bifunctional enzyme catalyzing both the hydrolysis of peptide bonds and transpeptidation. To achieve mature toxins, most of these resultant cyclic peptides undergo further posttranslational modifications, mainly including multiple hydroxylations, sulfoxidation, epimerization, and the formation of a cross-bridge between Trp and Cys; the order in which these reactions occur is unknown. The MSDIN genes encoding amatoxins, such as α- and β-amanitin, and phallotoxins, such as phallacidin and phalloidin, are readily found in the sequenced genomes of lethal Amanita species [15,18]. In addition, roughly 20–30 uncharacterized MSDIN genes have been identified in each of these genomes [18,19]. In the cases of Amanita bisporigera and A. phalloides, about half of the unknown MSDIN genes have been found to be expressed at the transcription level, and the results of mass spectrometry have indicated that two of the genes produce corresponding cyclic peptides [18,19]. In the past 10 years, only a few novel cyclic peptides have been discovered, including amanexitide in Amanita exitialis [20], and cycloamanides F and E (CylF and CylE) in A. phalloides [19]. In total, roughly 25 cyclic peptides have been described [1,21]. In contrast, newly sequenced genomes of lethal Amanita species show that the MSDIN genes by far outnumber the count of known cyclic peptides, indicating there could be a much larger potential for novel cyclic peptides in these mushrooms. However, it is mostly unknown whether these MSDINs are actually translated into cyclic peptides.
Conventional methods for cyclic peptide characterization in Amanita mainly use thin-layer chromatography (TLC), HPLC, hydrolysis, and NMR techniques, which generally need a significant amount of examined material [1,6]. Though somewhat biased due to monitoring wavelengths, HPLC analyses in many laboratories have suggested the most significant peptide production in Amanita species is attributed to the major toxins, i.e., α-amanitin, β-amanitin, phallacidin, and phalloidin. Noticeably, a number of less significant peaks are present in the vicinity of these known peptides, many of which are of insufficient amount for further analyses via conventional methods. Given the situation, a more sensitive method would be very appreciated.
Functionally speaking, cyclic peptides produced by Amanita species are more diverse than those commonly known from just the toxins. Antamanide, a cyclic decapeptide, protects mice from phalloidin toxicity. Through competition, the molecule effectively inhibits the uptake of phalloidin by the OATP1B1 (organic anion–transporting polypeptide family) transporter of the liver cells [22]; the same transporter is used by phallotoxins [23]. Newer findings suggest that the peptide is a novel inhibitor of the mitochondrial permeability transition pore [24]. Cycloamanides (CylA, CylB, CylC, and CylD), antamanide, and their synthetic derivatives display immunosuppressive activities in both in vivo and in vitro assays [25,26,27]. In A. phalloides, researchers suspect that CylE and CylF are related to cycloamanides due to sequence similarities [6]. These examples have placed many of the cyclic peptides outside the toxin category, indicating that they may have diverse functions in Amanita mushrooms.
In the present study, two lethal Amanita species, Amanita rimosa and A. exitialis, were sequenced, and their MSDIN genes mined. Together with two previous sequenced species, Amanita pallidorosea and Amanita subjunquillea, four sets of MSDIN genes were analyzed using a combination of methods. Linear and cyclic peptides were predicted by directly using amino acid sequences in the core regions of the MSDIN genes. Modifications after cyclization, such as hydroxylation, sulfoxidation, and cross-bridge formation, were taken into consideration. Possible molecular formulas with and without further modifications were generated for all candidate peptides. Through high resolution MS (HRMS), masses matching putative cyclic peptides were mined. Further amino acid composition was elucidated based on MS/MS results, which were manually analyzed via multiple peptide analyzing platforms. In addition, three synthetic peptides were obtained and included in this study for further structure confirmation.
2. Materials and Methods
2.1. Collection and Preservation of Samples
Fresh fruiting bodies of A. subjunquillea (As), A. exitialis (Ae), A. rimosa (Ar), and A. pallidorosea (Apa) were collected in the Yunnan, Guangdong, Hunan, and Shandong provinces of China, respectively. All the above samples were immediately submerged in dry ice on site, transferred to liquid nitrogen tanks the same day, freeze-dried, and then stored at −80 °C. All the mushrooms were taxonomically identified according to previous studies [3,5].
2.2. Genome and Transcriptome Sequencing
The sequencing platform for the genomes of Ae and Ar was PacBio Sequel at NextOmics Biosciences, Wuhan, China. Sequencing and assembly were carried out using the company’s standardized pipeline. High quality DNA was extracted as described before [18], and a 20-kbp library was constructed using a PacBio template prep kit and then analyzed by Agilent 2100 Bioanalyzer for quality control. After the completion of the library, the DNA template and enzyme complex were transferred to the Sequel sequencer for real-time single molecule sequencing. An Illumina HiSeq X10 platform was used for nucleotide level correction based on a 350-bp library and the company’s standard method. For the following analyses, two additional genomes of As and Apa were included from our previous studies [18].
One young fruiting body of As was chosen for transcriptome sequencing. The process was carried out via the standardized pipeline at NextOmics Biosciences, Wuhan, China. The library was constructed after the samples passed quality inspection. The complementary DNA (cDNA) library was constructed by magnetic bead enrichment. Clean reads were obtained from the Illumina RNA-Seq platform, these were then aligned to the As genome using the HISAT2 software [28]. Transcriptome assembly was performed using StringTie under default parameters [29].
2.3. Extraction of Cyclic Peptides
Freeze-dried material (0.05 g) for all samples was ground to fine powder with liquid nitrogen in a mortar and pestle. Then, a 1 mL extraction buffer containing absolute methanol:water:0.01 M hydrochloric acid (5:4:1) was added as described previously [30,31]. The mixture was heated in a water bath at 75 °C for 15 min before centrifugation at 12,000 rpm for 10 min. The supernatant was transferred to a new 1.5 mL centrifuge tube, filtered with a 0.22 μm polyethersulfone syringe filter (BS-PES-22, Biosharp), and then stored at −80 °C before use.
2.4. HPLC Analysis
An Agilent 1260 Infinity II HPLC system equipped with a UV detector (Agilent Technologies, Santa Clara, CA, USA) was employed for the analysis. Separations were carried out on an InfinityLab Poroshell 120 EC-C18 reverse phase HPLC column (4.6 × 100 mm I.D., particle size 2.7 μm, Agilent Technologies) at 28 °C. The mobile phases were (A) 0.02 M aqueous ammonium acetate–acetonitrile (90:10, v/v) and (B) 100% acetonitrile. The pH of solvent A was adjusted to 5.0 with glacial acetic acid. All solutions were prepared with HPLC-grade reagents and were degassed before use. For HPLC separation, the flow rate was 0.5 mL/min with constant 100% solution A for 2 min, followed by a gradient from 100% solution A to 100% solution B over an 8 min time course. The absorbance of the elution was monitored at 280 and 295 nm.
2.5. Mining MSDIN Genes
All nucleotide sequences of MSDINs from their genomes and the As transcriptome were obtained through standalone Basic Local Alignment Search Tool (BLAST) searches at National Center for Biotechnology Information (NCBI BLAST + 2.6.0) with query MSDIN sequences from Ab (EU196139–EU196158) and Galerina marginata [32], which are well-characterized by our molecular and biochemical approaches [15,32]. In addition, MSDINs with a high divergence at the core region from our previous reports were added as queries for a more thorough excavation of potential MSDIN genes [18].
2.6. Predicted Molecular Masses of Candidate Peptide Sequences
To obtain molecular masses of predicted peptides based on MSDIN sequences, the Molecular Isotopic Distribution Analysis (MIDAs) website (https://www.ncbi.nlm.nih.gov/CBBresearch/Yu/midas/index.html (accessed on 16 February 2021)) was used to calculate monoisotopic masses. This report mainly used the NCBI online calculation under the following conditions: coarse-grained isotopic distribution mass accuracy (Da) = 1; fine-grained isotopic distribution mass accuracy (Da) = 0.01; and the polynomial-based method. Linear peptides directly adopted the amino acid sequences for core regions of MSDIN genes. Cyclized versions equaled the linear versions minus a water molecule. Besides cyclization, this study attempted to predict other possible posttranslational modifications, including hydroxylation, sulfoxidation, and bridge formation. One hydroxylation adds one O atom; known amatoxins can contain as many as four hydroxylations. Sulfoxidation adds an O atom, which only happens to the peptides containing Cys and Trp. The bridge formation between Trp and Cys loses two H atoms, resulting in a structure called tryptathionine [33]. How many and in which order these modifications would occur to a given MSDIN-related peptide are unknown. Our strategy was to try different combinations and look for matches in LC–HRMS.
2.7. LC–MS and LC–MS/MS Analyses
HPLC separation, as described above, was coupled with Agilent 6540 UHD precision mass quadrupole time-of-flight (Q-TOF) LC/MS (Agilent Technologies) equipped with an electrospray source. Eluant was monitored in positive electrospray ionization (ESI) mode with the capillary voltage at 3.5 kV. The drying gas (N2) temperature was 350 °C, and the flow rate was 8 L/min. The mass scan range was 500–1700 m/z. For collision energy in the subsequent MS/MS analysis, a range of 10–70 eV was applied.
After LC–MS/MS data were obtained, amino acid composition and combinations of candidate cyclic peptides were investigated first through the platform at the Center for Computational Mass Spectrometry (CCMS) based on the UniProtKB/Swiss-Prot database (https://proteomics.ucsd.edu/ProteoSAFe/ (accessed on 16 February 2021)). The workflow used MS-GF + (V1.3.0) on the website by extracting masses of fragments using Agilent Mass Hunter v8.0.0 (Agilent Technologies). The fragmentation diagrams were saved as Materials and Geometry Format (MGF) files and uploaded to the CCMS [34]. When no results were obtained with the CCMS, other platforms were included in the MS/MS analysis including Mascot (with NCBIprot database), pNove v3.1 [35], and XCMS online [36]. MS Convert (http://proteowizard.sourceforge.net/tools.shtml (accessed on 16 February 2021)) was used to obtain suitable file formats for each application.
All results from the above online tools were further confirmed or corrected via a thorough manual pipeline. First, high quality MS/MS spectra were obtained by optimizing the energy in the Q-TOF process. Best spectra were selected by choosing the ones with most y-type ions found in the fragmentations of their corresponding linear peptides. In some cases, increased amounts of samples were applied for optimal MS/MS spectra. Common fragmentation patterns for linear peptides were used because our preliminary results suggested that they were useful. Then, y-type ions for each MS/MS spectrum were determined based on core peptides of MSDINs using Molecular Weight Calculator v6.50 (http://omics.pnl.gov (accessed on 16 February 2021)). Last, immonium ions were searched based on amino acid compositions. Only in situations in which most or all amino acids (by themselves or in the right combinations) in a given candidate cyclic peptide matched a MSDIN sequence did we deem the cyclic peptide real with the sequence designated by the core region of the MSDIN gene.
2.8. Cyclic Peptide Synthesis
All the predicted cyclic peptides found in this study were highly hydrophobic and difficult to chemically synthesize, but, fortunately, three of the cyclic peptides were successfully synthesized by Nanjing Peptide Biotech Ltd. (Nanjing, China). These synthesized cyclic peptides were dissolved in the same buffer as that used for extraction (absolute methanol:water:0.01 M hydrochloric acid = 5:4:1). The purity for each peptide was greater than 95% determined by HPLC and MALDI-TOF mass spectrometry. The MS/MS analysis described above was used to compare fragment ions from both natural and synthetic peptides.
3. Results
3.1. Amanita Genomes
Fruiting bodies of Ae and Ar were collected in excellent condition (Figure 1). For Ae, the N50 of the draft genome was 3.03 Mbp with a genome size of 49 Mbp (GC content = 46.6%), and the assembly contained 129 contigs. For Ar, the N50 was 690 kbp with the genome size at 45 Mbp (GC content = 45.99%), and the assembly comprised 134 contigs. The drafts for As and Ap were adopted from our previous report [18]. The Whole Genome Shotgun assemblies were deposited at DDBJ/ENA/GenBank as part of the accession JAEBUT000000000.
Figure 1.
Deadly Amanita species for genome sequencing and peptide analyses. (A) Amanita rimosa. (B) Amanita exitialis.
3.2. MSDIN Genes for Candidate Cyclic Peptides
With multiple known MSDINs as queries, the genomes revealed the presence of MSDIN genes in the two newly sequenced Amanita species, i.e., Ae and Ar, yielding 23 and 39 predicted MSDIN genes, respectively (Table 1 and Table 2). With expanded query MSDIN sequences, four additional MSDIN genes were found in As compared to our previous report (Table 3), but none were found in Ap. In total, 66 MSDINs were discovered, with 46 being novel. All genomes possessed MSDIN genes coding for at least three major cyclic peptide toxins, namely α-amanitin, β-amanitin, and phallacidin. Overall, the precursor genes share similar structures, designated as the leader peptide, core peptide, and recognition sequence [37]. Based on the newly discovered MSDINs, linear and cyclic peptides with and without further modifications were predicted. A schematic diagram of the genome-guided approach is shown in Figure 2.
Table 1.
The MSDIN gene family in Amanita rimosa.
| Leader Peptide | Core Peptide | Recognition Sequence | Monoisotopic Mass |
|---|---|---|---|
| MSDINSTRLP | IWGIGCNP | SVGDEVTALLTRGEA | 918.3541 (α-amanitin) |
| MSDINATRLP | IWGIGCNP | SVGDEVTALLASGEA | 918.3541 (α-amanitin) |
| MSDINATRLP | IWGIGCDP | CVGDDVAALTTRGEA | 919.3382 (β-amanitin) |
| MSDINATRVP | AWLVDCP | CVGDDISRLLTRGEK | 846.3217 (phallacidin) |
| MSDINATRLP | AWDSKHP | CVGDDVSRLLTRGE | 821.3820 |
| MSDINATRLP | AWDSKHP | CVGDDISRLLTRGE | 821.3820 |
| MSDINATRVP | AWLAECP | CVGDDISHLLTRGE | 770.3421 |
| MSDINASRLP | FFIIIVKP | CGNPYVSDDVNSTLTRGE | 957.6052 |
| MSDINTSRLP | FIPLGIITILP ★ | CVSDDVNTTITRGD | 1177.7475 |
| MSDINTACLP | FLFPVIPP | CLSEDANVVVLNSGE | 910.5317 |
| MSDINVTRLP | FFPIVFIPP | CI | 1057.6000 |
| MSDINIARLP | IFWFIYFP | CVGDDVDNTLSRGE | 1113.5688 |
| MSDINVTRLP | IFLIMFIPP | CIGDDAASILKQGE | 1071.6191 |
| MSDINTSCLP | IFIAFPIPP | CVSDDIQTVLTRGE | 995.5844 |
| MSDTNTACLP | IFIAFPIPP | CVSDDIQTVLTRGE | 995.5844 |
| MSDINASRLP | ILKKPWAP | SVCDDVNSTLTRGE | 933.5800 |
| MSDINVARLP | ISDPTAYP ★ | CVGDDIQAVVKRGE | 844.3967 |
| MSDINATRLP | IIIVLGLIIP | LCVSDIEMILTRGE | 1044.7311 |
| MSDINASRLP | IILAPIIP | CISDDVNTTLTCAE | 830.5630 |
| MSDINTTGLP | HFYNLMPP | CFSDDTGMVLVRGE | 999.4637 |
| MSDINATRLP | HPFPLGLQP | CAGDVDNFTLIKGE | 986.5338 |
| MSDINASCLP | LILVANGMAYV | --SDDVSPTLTRGE | 1144.6315 |
| MSDINTARLP | SYIPFPPP | CLSEDTNAVLMLGE | 898.4589 |
| MSDINTARLP | SYIPFPPP | CLSEDTNAVLMLGE | 898.4589 |
| MSDINTSRFP | SYGYRAFP | CVGDDVEMVLMHGE | 941.4396 |
| MSDINVTRLP | VLVFIFFLP | CISDDAASIIKLGE | 1075.6470 |
| MSDIDTTRLP | LILFTLQP | SIGDDVNPTLTRGEK | 925.5637 |
| MSDIHAARLP | FPTRPVFP ★ | SAGDDMIEVVLGRGE | 941.5123 |
| MSDNNAARLP | FYFYLGIP | SDDAHPILTRGERLA | 1000.5058 |
| MSDTNTARLP | ILFIQLEIP | CISDDVHPVLTRGE | 1066.6427 |
| MSDVNTTRLP | FNFFRFPYP | CICDDSEKVLELGE | 1215.5866 |
| MSEINTARFP | NHGHRTIP | CVGDDIEMVLMHGE | 912.4678 |
| MSEINTSRLP | LVFIPPYFAP | CVSDDIQMVLTLGE | 1144.6321 |
| MFDMNTTCLP | GFIIYAYV | --GDDVNHTLTRGE | 926.4902 |
| MLDINTARLP | FSLPTFPP | CVSDEIDVVLKRGE | 886.4589 |
| MLDINATRFP | LGRPTHLP | CVGDDVNYIL | 871.5028 |
| MTDINDARLP | ILLLIFFWIP | CANDDDENILNRG | 1255.7733 |
| MTDINDTRLP | FVWILWLWLA | CVGDDTSILNRGE | 1327.7481 |
| MPDINVTRLP | LLIIVLLTP | CISDDNNILNRGK | 975.6732 |
★ and underlined letters indicate novel cyclic peptides detected with MS and MS/MS. The monoisotopic masses are for unmodified cyclic peptides based on MSDIN core peptides, except for major toxins (α-amanitin, β-amanitin and phallacidin). Core peptides are indicated in bold.
Table 2.
The MSDIN gene family in Amanita exitialis.
| Leader Peptide | Core Peptide | Recognition Sequence | Monoisotopic Mass |
|---|---|---|---|
| MSDINATRLP | IWGIGCNP | CVGDDVTSVLTRGEA | 918.3541(α-amanitin) |
| MSDINATRLP | IWGIGCDP | CVGDDVTALLTRGEA | 919.3382(β-amanitin) |
| MSDINATRLP | AWLVDCP | CVGDDVNRLLTRGE | 846.3217(phallacidin) |
| MSDINATRLP | AWLTDCP | CVGDDVNRLLTRGE | 786.3371 |
| MSDINTTRLP | FVFVASPP ★ | CVGDDIAMVLTRGE | 844.4483 |
| MSDINTARLP | FIWVFGIP | --GDDIGTVLTRGEK | 959.5269 |
| MSDINLTRLP | GIIAIIP | CVGDDDDVNSTLTRGQ | 677.4476 |
| MSDINATRLP | IILAPVIP | CISDDNDP--TLTRGQ | 816.5473 |
| MSDINTARLP | IPIPPFFFP | FVSDDIEIVLRRGEK | 1055.5844 |
| MSDINTARLP | IPIPPFFFP | FVSDDIEIVLRRGEK | 1055.5844 |
| MSDINATRLP | IGRPQLLP | CVGGDVNYILISGEK | 874.5389 |
| MSDINPTRLP | IFWFIYFP | CVSDVDST-LTRGE | 1113.5688 |
| MSDINTARLP | IYRPPFYALP | CVGDDIQAVLTRGE | 1217.6597 |
| MSDINTARLP | IIWIIGNP | CVSDDVERILTRGE | 906.5327 |
| MSDINVIRAP | LLILSILP | CVGDDIEV-LRRGE | 862.5892 |
| MSDINATRLP | LFFPPDFRPP ★ | CVGDADNFTLTRGEK | 1213.6284 |
| MSDINATRLP | LFFPPDFRPP ★ | CVGDADNFTLTRGE | 1213.6284 |
| MSDINVIRLP | SMLTILPP | CVSDDASNTLTRGE | 852.4779 |
| MSDINTARLP | VFSLPVFFP ★ | --SDDIQAVLTRGE | 1033.5637 |
| MSDINVTRLP | VFIFFFIPP | CVGDGTADIVRKGEK | 1107.6157 |
| MSDINATRLP | VWIGYSP | CVGDDCIALLTRGE | 802.4014 |
| MSDINATRLP | VWIGYSP | CVGDDCIALLTRGE | 802.4014 |
| MTDINDTRLP | FIWLLWIWLP | SVGDD-NNILNRGEE | 1367.7794 |
★ and underlined letters show novel and known cyclic peptides detected with MS and MS/MS. The monoisotopic masses are for unmodified cyclic peptides based on MSDIN core peptides, except for major toxins (α-amanitin, β-amanitin and phallacidin). Core peptides are indicated in bold.
Table 3.
Expression of the MSDIN gene family from Amanita subjunquillea.
| Leader Peptide | Core Peptide | Recognition Sequence | Monoisotopic Mass | Expressed |
|---|---|---|---|---|
| MSDINATCLP | IWGIGCNP | CVGDEVAALLTRGEALC | 918.3541(α-amanitin) | √ |
| MSDINATRLP | IWGIGCDP | CVGDEVTALLTRGEALC | 919.3382 (β-amanitin) | √ |
| MSDINATRLP | IWGIGCDP | CIGDDVTALLTRGEALC | 919.3382 (β-amanitin) | √ |
| MSDINATRLP | AWLATCP | CAGDDVNPTLTRGESLC | 788.3160 (phalloidin) | √ |
| MSDINATRLP | AWLVDCP | CVGDDINRRVVSAFA-C | 846.3217 (phallacidin) | √ |
| MSDMNATRLP | LIQRPFAP | CVSDDVDFALIRRCALVYAESSV | 922.5389 | √ |
| MSDINTARLP | HFASFIPP | CIGDDIEMVLKRGESLC | 896.4545 | √ |
| MSDINTARLP | TFLPPLFVPP ★ | CVSDDIEMVLTRGESLC | 1108.6321 | √ |
| MSDINATRLP | LNILPFMLPP | CVGDDVNPTLTRGEDLC | 1135.6464 | √ |
| MSDMNATRLP | LIQRPYAP | CVSDDVNSPLTRGESLC | 938.5338 | √ |
| MSDINTARLP | IGRPESIP | CVGDDIEMILERGQKLC | 849.4709 | √ |
| MSDINTARLP | LRLPPFMIPP | CVGDDIGMVLTRGENLC | 1161.6733 | √ |
| MSDVNATRLP | FNFFRFPYP | CIGDDSASVLGLGESLC | 1215.5866 | √ |
| MSDINATRLP | SSVLPRP | CVGDVDNIILTSREKLC | 736.4232 | √ |
| MSDINTARLP | AFFPPFFIPP ★ | CVSDDIEMVLTRGESLC | 1160.6059 | √ |
| MSDINATRLP | IPILPIPP | YCSDDANTTLTLGESLC | 840.5473 | √ |
| MSDINATRLP | LFLLAALGIP | --SDDADSTLTRGESLC | 1008.6372 | √ |
| MSDTNDARLP | LFFWFWFLWP | SVSDDIDSVLNRGEDLC | 1469.7325 | √ |
| MSDMNVARLP | ISDPTAYP | CVGGDIHAVLRRGE | 844.3966 | × |
| MSDMNVARLP | ISDPTAYP | CVGGDIHAVLRRGE | 844.3966 | × |
| MSDINVTCLP | FIFWFFWPP | CVGDDAASIIK-GK | 1267.6218 | × |
| MSDINAARLP | FIFPPFFIPP | CVSDDIEMVLTRGE | 1202.6528 | × |
| MSDINTVCLP | LQKPWSRP | CVGDDIEMILERGE | 992.5556 | × |
| MFDINITRLP | IFWFIYFP | CVGDDVTALLTRGE | 1113.5689 | × |
★ and underlined letters indicate novel cyclic peptides detected with MS and MS/MS. The monoisotopic masses are for unmodified cyclic peptides based on MSDIN core peptides, except for major toxins (α-amanitin, β-amanitin, phalloidin and phallacidin). Grey letters indicate alternative splicing. Core peptides are indicated in bold.
Figure 2.
A flowchart for characterizing cyclic peptides in Amanita species. HRMS: high resolution MS.
3.3. Transcriptome of As
As was chosen to be sequenced via the Illumina RNA-Seq technique, as it was a readily available species. In total, 11.7 Gb clean reads were obtained. The assembled transcriptome yielded 46.1 Mbp with 25,453 unigenes. A BLAST search against the transcriptome with MSDINs from the As genome produced 22 MSDINs (Table 3), four more than those in our previous result [18]. These four sequences were used as queries to re-search the genome, and the result confirmed their presence. Eighteen out of the 22 MSDIN genes were found to be expressed at the transcription level, i.e., 81.8% of the MSDINs were expressed (Table 3). Regarding the gene structure, exons and introns were conserved among most of the MSDIN genes, while alternative splicing was detected in two of the MSDIN transcripts marked in grey in Table 3. The expressed MSDINs were the focus for potential cyclic peptide production in the following MS and MS/MS analyses.
3.4. LC–HRMS and LC–MS/MS Analyses of Novel Cyclic Peptides in As
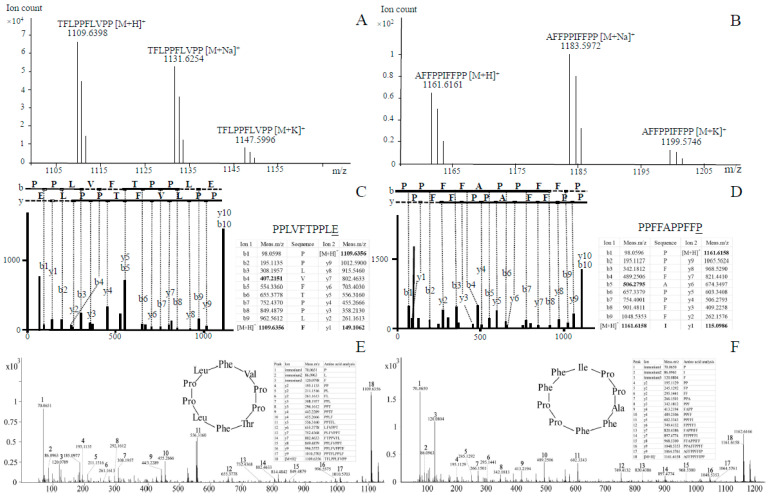
With the Agilent LC–HRMS platform, correlations between measured masses and predicted peptides based on genomic data were carefully assessed. No linear versions of these peptides were detected. Further, hydroxylation(s), sulfoxidation, and cross-bridging (by themselves or by various combinations) were not detected on predicted linear or cyclic peptides. However, two matches were found between theoretical and measured masses of two predicted cyclic peptides with no further modifications. These two matches corresponded to the cyclized core peptide sequences of TFLPPLFVPP (designated CylG1) and AFFPPFFIPP (named CylG2), respectively. The molecular formula for the candidate new cyclic peptide CylG1 is C59H84N10O11, and the theoretical molecular weight was found to be 1109.6394 [M+H]+. The measured molecular weight was 1109.6398 [M+H]+, with a mass discrepancy of 0.36 ppm (Figure 3A). The molecular formula for CylG2 is C65H80N10O10 with the theoretical molecular weight at 1161.6132 [M+H]+; the measured molecular weight was 1161.6161 [M+H]+ with a mass discrepancy of 2.50 ppm (Figure 3B). Two adduct ions of [M+Na]+ and [M+K]+ are also shown in the figure. CylG1 and CylG2 were treated as candidate new cyclic peptides for further characterization.
Figure 3.
LC–HRMS and the amino acid composition of the cyclized core peptide sequences of TFLPPLFVPP and AFFPPFFIPP (CylG1 and CylG2, respectively) in Amanita subjunquillea. Positive LC–HRMS spectra of CylG1 (A) and CylG2 (B), with [M+Na]+ and [M+K]+ adduct ions indicated. (C,D) Amino acid composition analysis through the Center for Computational Mass Spectrometry (CCMS) for CylG1 and CylG2, with b- and y-type fragment ions shown (bold fonts were manually added for ions not recognized by the website); predicted linear peptide sequences are shown above the inserted tables (the last amino acids are underlined as they were erroneous). (E,F) Manual analyses of recognized fragment ions for CylG1 and CylG2 with inserted tables of matched amino acids; immonium ions are shown when present. The determined cyclic peptides are shown as circles.
In order to determine the amino acid composition of CylG1 and CylG2, the candidate cyclic peptides were submitted to LC–MS/MS. The resulting spectra were first analyzed by the CCMS using the UniProtKB/Swiss-Prot database (Figure 3C). The calculated peptide sequence for CylG1 was PPLVFTPPLE. The amino acid composition differed at one position from the original sequence (E vs. F), which could be attributed to the application of a linear peptide database in the CCMS (Figure 3C underlined). The molecular weight of F was found to be 165.19, and the loss of water during cyclization resulted in a mass of 147.18, which was in agreement with the calculated mass of E (147.13) and therefore accounted for the presence of E over F. For Cy1G2, similar processes were applied, and a discrepancy was also found at the last predicted amino acid (underlined in Figure 3D). Some of our peptides did not return any results with the CCMS, and they were then analyzed with other platforms (Mascot, pNove, and XCMS). In general, the results were largely in line with the above. Though the automated pipelines offered some evidence, we mostly relied on the following manual process to determine the peptide sequences.
Manual amino acid composition analysis was mostly based on the y-type fragmentation of linear peptides. For CylG1, fragment ions were calculated using the core peptide, and the result of searching for these ions in the MS/MS spectrum is shown in Figure 3E. Every fragment ion was manually checked and confirmed. As illustrated in Figure 3E, y-type fragments (y2–y9) were found to be in strong agreement with the core peptide, and all the amino acids could be readily explained by the y-type ions. Immonium ions for P, L, and F were also identified for CylG1. As a result, CylG1 was assigned as a novel cyclic peptide with the amino acid composition and combinations matching the circular MSDIN core peptide sequence Cyclo (TFLPPLFVPP) (Figure 3C,E). CylG2 underwent the same analyses, in which case immonium ions for P, I, and F were identified. Similarly, y-type fragments (y2–y9) were shown to be in strong agreement with the core peptide, and all the amino acids complied with the y-type fragment ions (Figure 3D,F). In conclusion, CylG2 was confirmed to be a novel cyclic peptide with the sequence Cyclo (AFFPPFFIPP). The circular peptide sequence of CylG2 was found to be highly similar to antamanide, Cyclo (FFVPPAFFPP), differing by one amino acid (I vs. V).
3.5. Novel Cyclic Peptides in Other Amanita Species
CylG1 and CylG2 in As comprised our first effort towards finding novel cyclic peptides in the genus. Taking advantage of this genome-guided approach, we analyzed three other Amanita species, i.e., Apa, Ae, and Ar. In Ar, three new cyclic peptides with the sequences Cyclo (ISDPTAYP), Cyclo (FIPLGIITILP), and Cyclo (FPTRPVFP) were found (Supplementary Figures S1 and S2), which were named CylH1, CylH2, and CylH3, respectively. In Apa, five new cyclic peptides with the sequences Cyclo (EFIVFGIFP), Cyclo (FVIIPPFIFP), Cyclo (YFFNDHPP), Cyclo (TIHLFSAP), and Cyclo (MHILAPPP) (Supplementary Figures S3 and S4), which were named CylI1, CylI2, CylI3, CylI4, and CylI5, respectively. Further information on MSDIN genes in Apa are included in Supplementary Table S1. In Ae, two new cyclic peptides with the sequences Cyclo (FVFVASPP) and Cyclo (LFFPPDFRPP) were found (Supplementary Figures S5 and S6), which were named CylJ1 and CylJ2, respectively. A previously known cyclic peptide called amanexitide with the sequence Cyclo (VFSLPVFF) [20] was also found in A. exitialis in this study (Supplementary Figure S7). In conclusion, 12 novel cyclic peptides and one known cyclic peptide were discovered in the four sequenced species (Table 4). Relevant data analyses of mass spectrometry are included in Supplementary Figures S1–S7, Supplementary Table S1.
Table 4.
Twelve novel cyclic peptides and one known cyclic peptide discovered in Amanita subjunquillea, A. rimosa, A. exitialis, and Amanita pallidorosea.
| Species | Cyclopeptide Sequence | Molecular Formula | Theoretical (m/z) | Measured (m/z) | δ (ppm) |
|---|---|---|---|---|---|
| A. subjunquillea | TFLPPLFVPP (CylG1) | C59H84N10O11 | 1109.6394 | 1109.6398 | 0.36 |
| AFFPPFFIPP (CylG2) | C65H80N10O10 | 1161.6132 | 1161.6161 | 2.50 | |
| A. rimosa | ISDPTAYP * (CylH1) | C39H56N8O13 | 845.4039 | 845.4040 | 0.12 |
| FIPLGIITILP (CylH2) | C61H99N11O12 | 1178.7547 | 1178.7555 | 0.68 | |
| FPTRPVFP (CylH3) | C48H67N11O9 | 942.5196 | 942.5191 | 0.53 | |
| A. pallidorosea | EFIVFGIFP (CylI1) | C56H75N9O11 | 1050.5658 | 1050.5694 | 3.43 |
| FVIIPPFIFP (CylI2) | C65H90N10O10 | 1171.6914 | 1171.6941 | 2.31 | |
| YFFNDHPP (CylI3) | C51H59N11O12 | 1018.4417 | 1018.4421 | 0.39 | |
| TIHLFSAP (CylI4) | C42H62N10O10 | 867.4723 | 867.4733 | 1.15 | |
| MHILAPPP (CylI5) | C41H64N10O8S | 857.4702 | 857.4714 | 1.40 | |
| A. exitialis | FVFVASPP (CylJ1) | C44H60N8O9 | 845.4556 | 845.4582 | 3.08 |
| LFFPPDFRPP # (CylJ2) | C63H83N13O12 | 1214.6357 | 1214.6357 | 0.00 | |
| VFSLPVFFP ^ | C56H75N9O10 | 1034.5709 | 1034.5734 | 2.42 |
* indicates that the sequence has been found in A. subjunquillea, A. pallidorosea, and A. rimosa. # indicates this sequence has two copies. ^ indicates amanexitide.
3.6. Confirmation of Three Cyclic Peptides via Synthetic Cyclic Peptides
Two of the novel cyclic peptides (CylG1 and CylG2) and amanexitide were chemically synthesized, and MS/MS data were obtained. The majority of fragment ions were shared between the natural and synthetic peptides in each pair (Figure 4). For CylG1, 16 primary fragment ions were consistent in both natural and synthetic compounds; for CylG2, 18 primary fragment ions were identical; and for amanexitide, 24 fragment ions were matched.
Figure 4.
MS/MS fragment comparison of three natural and synthetic cyclic peptides from Amanita species. Fragment ions of natural and synthetic CylG1 (A), CylG2 (B), and amanexitide (C). CylG1 and CylG2 are produced by A. subjunquillea, and amanexitide is produced by A. exitialis. Shared ions are labeled.
4. Discussion
Edible Amanita species are consumed in many parts of the world purposefully and by accident [6]. In the local markets of Yunnan Province, China, we constantly find at least three edible Amanita species. Unfortunately, many lethal Amanita species share morphological characteristics with the edible ones, leading to misidentification, consumption, and serious poisonings. In 2019, the Chinese Center for Disease Control and Prevention (China CDC) reported 276 independent mushroom poisonings in China with 769 patients and 22 deaths, and the majority of the deaths (20 cases) were caused by Amanita and amanitin-containing species [38]. The genome-guided approach adopted in this study identified 12 novel cyclic peptides and one known cyclic peptide in four Amanita species. The reliability of the results was furthered confirmed by comparing three of the natural peptides with synthetic ones. Our results indicated that there is a large cyclic peptide reservoir in Amanita species and provides a fast method of detection.
Traditional methods for identifying novel cyclic peptides in lethal Amanita species rely heavily on the availability of sufficient examined materials [1,6]. The MS-based method adopted in this study is much more sensitive, requiring only good MS/MS spectra. The first report using MS (as part of the approach) for identifying novel cyclic peptides in these mushrooms showed some initial promise [20], but the lack of genomic information limited its further application. An automated MS-based pipeline indicated that there are likely two novel cyclic peptides in A. phalloides [19], which pointed out a new direction, although manual analysis and further confirmation were lacking. In this study, manual analyses of MS and MS/MS data were shown to be the key for the pipeline. It is also worth noting that the MS analysis of the cyclic peptides required good sample preparation and separation. Obtaining optimal MS/MS spectra oftentimes needed tweaking with collision energy and sample amounts. Fragment ions could frequently only be recognized by manual inspection.
Most of the MSDIN genes were expressed at the transcription level in As, as suggested by the RNA-Seq data. Previous studies detected as many as half of MSDINs expressed in A. phalloides and A. bisporigera [19], and additional expressed sequences were cloned from other Amanita species through RT-PCR [39,40]. These data indicated MSDINs are mostly real, active genes that could potentially lead to production of many more cyclic peptides than previously known in the genus. Our results also strongly suggest that the amanitin biosynthetic pathway is highly versatile and capable of producing a wide range of cyclic peptides.
The cyclic peptides, including the toxins, are highly valuable resources. To date, most cyclic peptides in Amanita were first discovered from A. phalloides with a few exceptions [6]. It has been suspected that lethal Amanita species could produce significantly more cyclic peptides than what have been discovered [1,6]. The cyclic peptides discovered in this study lack further modifications besides cyclization and could therefore be readily synthesized for the purpose of screening for useful activities. Cyclic peptide pools in different species have distinctions, although the core set (major toxins) is relatively stable. As a result, the collection of the peptides in dozens of known lethal species in the genus should constitute a large cyclic peptide reservoir, potentially valuable for useful biological activities. α-Amanitin has been shown to have high potential in cancer research and treatment [41,42]. Phalloidin and phallacidin are valuable in cell research as generic molecular tools [43]. Antamanide functions as an antitoxin, reversing the effort of phalloidin [44], and researchers now know that antamanide inhibits the mitochondrial permeability transition pore by targeting the regulator cyclophilin D [24]. In addition, cycloamanides and related peptides possess potent activities useful in immunity-related medical situations [25,27]. Consequently, unknown cyclic peptides in Amanita species have the potential to bear useful activities.
Regarding the structures of the novel cyclic peptides, CylG1 and CylG2 have a characteristic amino acid composition with double Pro residues, no Trp and Cys, and without any further posttranslational modifications. These compounds and amanexitide share similarities to antamanide in that they contain ten amino acids (decapeptides) with a conserved sequence of XXXPPXXXPP; the two pairs of Pro residues are frequently found in MSDINs. According to the MS/MS data, hydroxylation on the 12 evaluated peptides was not detected. Pro (100%), Phe (84.6%), and Ile (53.8%) were the most frequent residues in these cyclic peptides. CylG2 is very similar to antamanide, with only one amino acid difference (I vs. V). In Ar, the new peptide Cyclo (FIPLGIITILP), CylH2, has 11 amino acids, which is by far the longest in all discovered Amanita cyclic peptides. Despite some structural similarities, the biological activities of these peptides require further study.
5. Conclusions
In this report, two deadly Amanita species (A. rimosa and A. exitialis) were sequenced, and, in total, 46 novel MSDIN genes discovered in four draft genomes. A majority of the MSDIN genes were found to be expressed at the mRNA level in A. subjunquillea. Using mass spectrometry, 12 novel cyclic peptides, which are potentially valuable for useful activities, were identified. The results were further confirmed by comparing three natural peptides with synthetic ones. In conclusion, the results suggest that the genome-guided approach is robust for identifying novel cyclic peptides in lethal Amanita species.
Acknowledgments
We thank the Yunnan Key Laboratory for Fungal Diversity and Green Development, Kunming, Yunnan, Scientific Research Foundation of the Education Department of Human Resources and Social Security of Yunnan Province, China, and Scientific Research Foundation of Kunming Institute of Botany, Chinese Academy of Sciences. We have profound gratitude to Jonathan D. Walton for his very kind assistance for the past 12 years, and we are terribly sorry for his recent passing. We also thank Gengshen Wang and Y.L. for their kind help on the initial version of the manuscript. In addition, we thank Tai-Hui Li and Wang-qiu Deng for helping collect fruiting bodies of A. exitialis.
Supplementary Materials
The following are available online at https://www.mdpi.com/2309-608X/7/3/204/s1. Figure S1: The monoisotopic masses of candidate new cyclic peptides in Amanita rimosa. Figure S2: Tandem mass spectrometry of novel cyclic peptides in Amanita rimosa. Figure S3: The monoisotopic masses of candidate new cyclic peptides in Amanita pallidorosea. Figure S4: Tandem mass spectrometry of novel cyclic peptides in Amanita pallidorosea. Figure S5: The monoisotopic masses of candidate new cyclic peptide in Amanita exitialis. Figure S6: Tandem mass spectrometry of novel cyclic peptides in Amanita exitialis. Figure S7: Mass spectrometry of amanexitide from Amanita exitialis. Table S1: MSDIN family and novel cyclic peptides in Amanita pallidorosea.
Author Contributions
H.L. and S.Z. designed and carried out the genome sequencing, LC–MS/MS, and the analyses on fragment ions; S.Z. did the online database analysis. X.L. (Xincan Li), Y.L., and X.L. (Xuan Li) assembled the transcriptome and helped write the manuscript. Z.H.C. and G.L. collected the samples. H.L. and P.Y. assembled and analyzed the transcriptome. Z.L.Y. helped design the study and helped improve the final manuscript. H.L. and S.Z. prepared the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant no. XDB31000000), National Natural Science Foundation of China (Grant no. 31772377 and 31972477).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The Whole Genome Shotgun assemblies were deposited at DDBJ/ENA/GenBank as part of the accession JAE-BUT000000000.
Conflicts of Interest
All authors declared to have no conflict of interest.
Footnotes
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Wieland T. Peptides of Poisonous Amanita Mushrooms. Springer; Berlin/Heidelberg, Germany: 1986. [Google Scholar]
- 2.Bresinsky A., Besl H. A Colour Atlas of Poisonous Fungi: A Handbook for Pharmacists, Doctors, and Biologists. CRC Press; London, UK: 1990. [Google Scholar]
- 3.Yang Z.L. Atlas of the Chinese Species of Amanitaceae. Science Press; Beijing, China: 2015. [Google Scholar]
- 4.Chen Z.H., Yang Z.L., Bau T., Li T.H. Poisonous Mushrooms: Recognition and Poisoning Treatment. Science Press; Beijing, China: 2016. [Google Scholar]
- 5.Cui Y.Y., Cai Q., Tang L.P., Liu J.W., Yang Z.L. The family Amanitaceae: Molecular phylogeny, higher-rank taxonomy and the species in China. Fungal Divers. 2018;91:5–230. doi: 10.1007/s13225-018-0405-9. [DOI] [Google Scholar]
- 6.Walton J.D. The Cyclic Peptide Toxins of Amanita and Other Poisonous Mushrooms. Springer International Publishing AG; Cham, Switzerland: 2018. [Google Scholar]
- 7.Lynen F., Wieland T. Über die Giftstoffe des Knollenblätterpilzes. IV. Liebigs Ann. Chem. 1938;533:93–117. doi: 10.1002/jlac.19385330105. [DOI] [Google Scholar]
- 8.Wieland T., Dudensing C. Über die Giftstoffe des grünen Knollenblätterpilzes, XI. γ-Amanitin, eine weitere Giftkomponente. Justus Liebigs Ann. Chem. 1941;600:156–160. doi: 10.1002/jlac.19566000211. [DOI] [Google Scholar]
- 9.Diaz J.H. Evolving global epidemiology, syndromic classification, general management, and prevention of unknown mushroom poisonings. Crit. Care Med. 2005;33:419–426. doi: 10.1097/01.CCM.0000153530.32162.B7. [DOI] [PubMed] [Google Scholar]
- 10.Bushnell D.A., Patrick C., Kornberg R.D. Structural basis of transcription: α-Amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc. Natl. Acad. Sci. USA. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Greenleaf A.L.B., Leslie M., Jiamachello P.F., Coulter D.E. α-Amanitin-resistant D. melanogaster with an altered RNA polymerase II. Cell. 1979;18:613–622. doi: 10.1016/0092-8674(79)90116-8. [DOI] [PubMed] [Google Scholar]
- 12.Wieland T. The Toxic Peptides of Amanita Phalloides. Springer; Vienna, Austria: 1967. [DOI] [PubMed] [Google Scholar]
- 13.Wieland T., Schnabel H.W. Über die Giftstoffe des grünen Knollenblätterpilzes, XXI: Die Konstitution des Phallacidins. Liebigs Ann. Chem. 1962;657:218–225. doi: 10.1002/jlac.19626570127. [DOI] [Google Scholar]
- 14.Wieland T., Schnabel H.W. Über die Giftstoffe des grünen Knollenblätterpilzes, XXII: Neue Sequenzanalyse von Phalloidin und Phalloin. Liebigs Ann. Chem. 1962;657:225–228. doi: 10.1002/jlac.19626570128. [DOI] [Google Scholar]
- 15.Hallen H.E., Luo H., Scott-Craig J.S., Walton J.D. Gene family encoding the major toxins of lethal Amanita mushrooms. Proc. Natl. Acad. Sci. USA. 2007;104:19097–19101. doi: 10.1073/pnas.0707340104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo H., Hong S.Y., Sgambelluri R.M., Angelos E., Li X., Walton J.D. Peptide macrocyclization catalyzed by a prolyl oligopeptidase involved in α-amanitin biosynthesis. Chem. Biol. 2014;21:1610–1617. doi: 10.1016/j.chembiol.2014.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sgambelluri R.M., Smith M.O., Walton J.D. Versatility of prolyl oligopeptidase B in peptide macrocyclization. ACS Synth. Biol. 2018;7:145–152. doi: 10.1021/acssynbio.7b00264. [DOI] [PubMed] [Google Scholar]
- 18.Luo H., Cai Q., Lüli Y.J., Li X., Sinha R., Hallen–Adams H.E., Yang Z.L. The MSDIN family in amanitin-producing mushrooms and evolution of the prolyl oligopeptidase genes. IMA Fungus. 2018;9:225–242. doi: 10.5598/imafungus.2018.09.02.01. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pulman J.A., Childs K.L., Sgambelluri R.M., Walton J.D. Expansion and diversification of the MSDIN family of cyclic peptide genes in the poisonous agarics Amanita phalloides and A. bisporigera. BMC Genom. 2016;17:1038. doi: 10.1186/s12864-016-3378-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xue J.H., Wu P., Chi Y.L., Xu L.X., Wei X.Y. Cyclopeptides from Amanita exitialis. Nat. Prod. Bioprospect. 2011;1:52–56. doi: 10.1007/s13659-011-0013-9. [DOI] [Google Scholar]
- 21.Clarke D.B., Lloyd A.S., Robb P. Application of liquid chromatography coupled to time-of-flight mass spectrometry separation for rapid assessment of toxins in Amanita mushrooms. Anal. Methods. 2012;4:1298–1309. doi: 10.1039/c2ay05575a. [DOI] [Google Scholar]
- 22.Letschert K., Faulstich H., Keller D., Keppler D. Molecular characterization and inhibition of amanitin uptake into human hepatocytes. Toxicol. Sci. 2006;91:140–149. doi: 10.1093/toxsci/kfj141. [DOI] [PubMed] [Google Scholar]
- 23.Meier-Abt F., Faulstich H., Hagenbuch B. Identification of phalloidin uptake systems of rat and human liver. Biochim. Biophys. Acta Biomembr. 2004;1664:64–69. doi: 10.1016/j.bbamem.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 24.Azzolin L., Antolini N., Calderan A., Ruzza P., Sciacovelli M., Marin O., Mammi S., Bernardi P., Rasola A. Antamanide, a derivative of Amanita phalloides, is a novel inhibitor of the mitochondrial permeability transition pore. PLoS ONE. 2011;6:e16280. doi: 10.1371/journal.pone.0016280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Siemion I.Z., Pedyczak A., Trojnar J., Zimecki M., Wieczorek Z. Immunosuppressive activity of antamanide and some of its analogs. Peptides. 1992;13:1233–1237. doi: 10.1016/0196-9781(92)90034-Z. [DOI] [PubMed] [Google Scholar]
- 26.Wieczorek Z., Siemion I.Z., Zimecki M., Bolewska-pedyczak E., Wieland T. Immunosuppressive activity in the series of cycloamanide peptides from mushrooms. Peptides. 1993;14:1–5. doi: 10.1016/0196-9781(93)90003-Y. [DOI] [PubMed] [Google Scholar]
- 27.Thell K., Hellinger R., Schabbauer G., Gruber C.W. Immunosuppressive peptides and their therapeutic applications. Drug Discov. Today. 2014;19:645–653. doi: 10.1016/j.drudis.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim D., Landmead B., Salzberg S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea M., Pertea G.M., Antonescu C.M., Chang T.C., Mendell J.T., Salzberg S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hallen H.E., Watling R., Adams G.C. Taxonomy and toxicity of Conocybe lactea and related species. Mycol. Res. 2003;107:969–979. doi: 10.1017/S0953756203008190. [DOI] [PubMed] [Google Scholar]
- 31.Enjalbert F., Gallion C., Jehl F., Monteil H., Faulstich H. Simultaneous assay for amatoxins and phallotoxins in Amanita phalloides Fr. by high-performance liquid chromatography. J. Chromatogr. 1992;598:227–236. doi: 10.1016/0021-9673(92)85052-U. [DOI] [PubMed] [Google Scholar]
- 32.Luo H., Hallen-Adams H.E., Scott-Craig J.S., Walton J.D. Ribosomal biosynthesis of α-amanitin in Galerina marginata. Fungal Genet. Biol. 2012;49:123–129. doi: 10.1016/j.fgb.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.May J.P., Perrin D.M. Tryptathionine bridges in peptide synthesis. Biopolymers. 2007;88:714–724. doi: 10.1002/bip.20807. [DOI] [PubMed] [Google Scholar]
- 34.Kim S., Pevzner P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014;5:5277. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang H., Chi H., Zhou W.J., Zeng W.F., He K., Liu C., Sun R.X., He S.M. Open-pNovo: De novo peptide sequencing with thousands of protein modifications. J. Proteome Res. 2017;16:645–654. doi: 10.1021/acs.jproteome.6b00716. [DOI] [PubMed] [Google Scholar]
- 36.Tautenhahn R., Patti G.J., Rinehart D., Siuzdak G. XCMS Online: A web-based platform to process untargeted metabolomic data. Anal. Chem. 2012;84:5035–5039. doi: 10.1021/ac300698c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Arnison P.G., Bibb M.J., Bierbaum G., Bowers A.A., Bugni T.S., Bulaj G., Camarero J.A., Campopiano D.J., Challis G.L., Clardy J., et al. Ribosomally synthesized and post-translationally modified peptide natural products: Overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 2013;30:108–160. doi: 10.1039/C2NP20085F. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H.J., Zhang H.S., Zhang Y.Z., Zhang K.P., Sun C.Y. Mushroom poisoning outbreaks—China, 2019. China CDC Wkly. 2020;2:19–24. doi: 10.46234/ccdcw2020.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li P., Deng W.Q., Li T.H., Song B., Shen Y.H. Illumina-based de novo transcriptome sequencing and analysis of Amanita exitialis basidiocarps. Gene. 2013;532:63–71. doi: 10.1016/j.gene.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 40.Li P., Deng W.Q., Li T.H. The molecular diversity of toxin gene families in lethal Amanita mushrooms. Toxicon. 2014;83:59–68. doi: 10.1016/j.toxicon.2014.02.020. [DOI] [PubMed] [Google Scholar]
- 41.Liu Y., Zhang X., Han C., Wan G., Huang X., Ivan C., Jiang D., Rodriguez-Aguayo C., Lopez-Berestein G., Rao P.H., et al. TP53 loss creates therapeutic vulnerability in colorectal cancer. Nature. 2015;520:697–701. doi: 10.1038/nature14418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kume K., Ikeda M., Miura S., Ito K., Sato K.A., Ohmori Y., Endo F., Katagiri H., Ishida K., Ito C., et al. α-Amanitin restrains cancer relapse from drug-tolerant cell subpopulations via TAF15. Sci. Rep. 2016;6:25895. doi: 10.1038/srep25895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Craig E.W., Avasthi P. Visualizing filamentous actin using phalloidin in chlamydomonas reinhardtii. Bio Protocol. 2019;9:e3274. doi: 10.21769/BioProtoc.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raymond G., Potreau D., Cognard C., Jahn W., Wieland T. Antamanide antagonizes the phalloidin-induced negative inotropic effect and blocks voltage dependently the fast outward K+ current in voltage-clamped frog muscle fibres. Eur. J. Pharmacol. 1987;138:21–27. doi: 10.1016/0014-2999(87)90332-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Whole Genome Shotgun assemblies were deposited at DDBJ/ENA/GenBank as part of the accession JAE-BUT000000000.