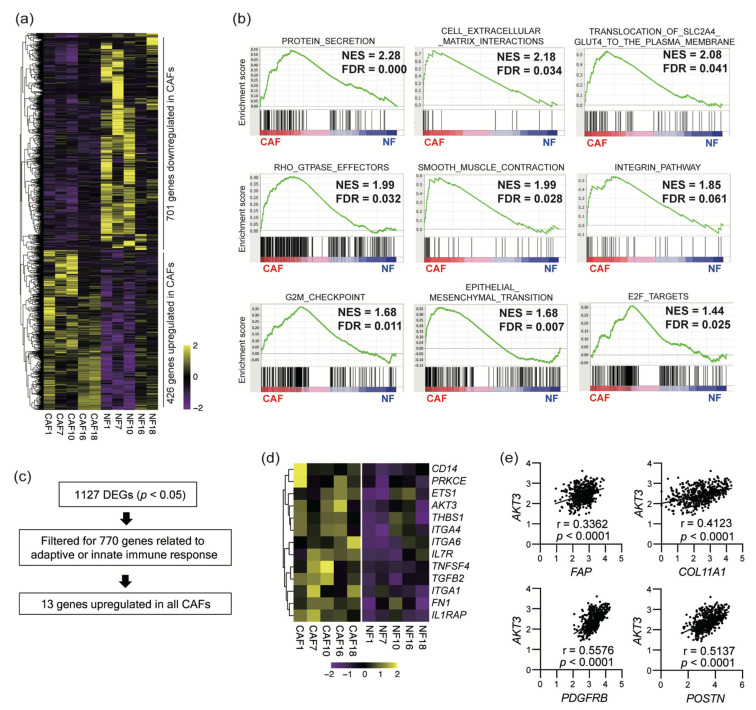
Figure 2.
RNA sequencing revealed a CAF-specific gene signature that included immune-related genes. (a) Heat map of differentially expressed genes (DEGs) in 5 paired CAF (cancer-associated fibroblast) and NF (normal fibroblast) samples obtained by RNA sequencing (p < 0.05). (b) Representative gene set enrichment analysis (GSEA) plots of hallmark gene sets and C2 canonical pathways comparing CAF-specific DEGs (FDR < 0.05). (c) A total of 1127 DEGs were searched for entries on a list of 770 genes that regulate adaptive or innate immune functions. Thirteen genes were found. (d) Heat map of 13 genes that were extracted from the 1127 DEGs in (d). (e) Correlation between the normalized expression of AKT3 and that of CAF-specific markers that were overexpressed in HNSCC tissues. mRNA expression data was obtained from TCGA.