Fig. 1.
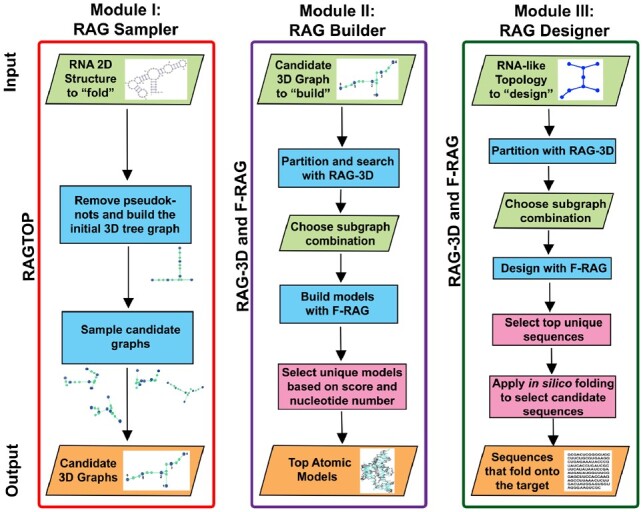
Flowchart for the three webserver modules. RAG Sampler takes RNA sequence and 2D structure as input, and generates corresponding candidate tree graph topologies. RAG Builder takes RNA sequence, 2D structure and target tree graph topology (generated by RAG Sampler) as input and builds corresponding atomic models. RAG Designer takes a target tree graph topology as input, and designs sequences that fold onto target topologies. The input and output information for the three modules is listed in the first and the last rows, respectively. Steps that involve running the three standalone tools (RAGTOP, RAG-3D, and F-RAG) and subsequent result analysis is indicated in rectangular boxes. Parallelograms are used for steps where user input is required during processing
