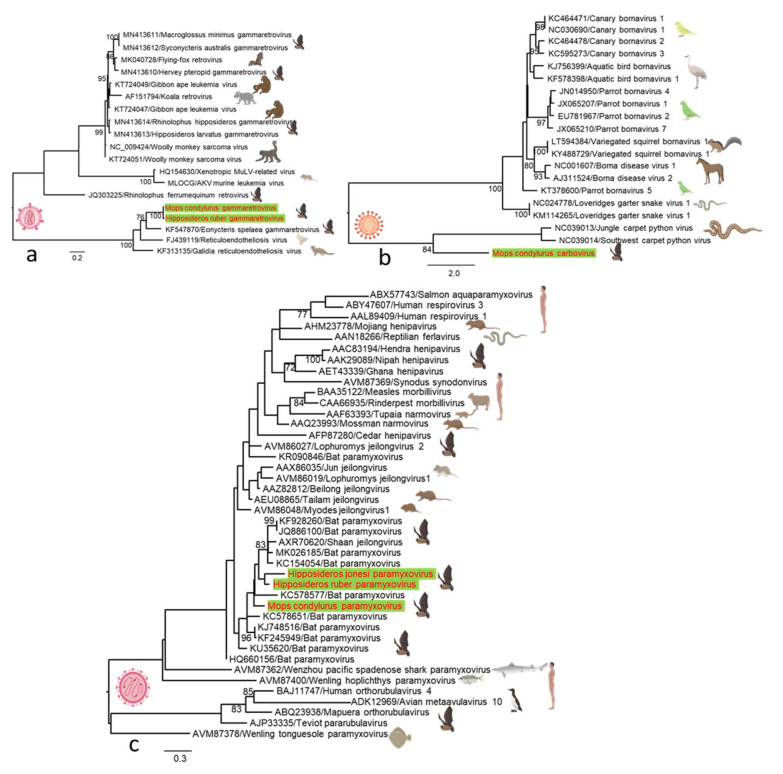
Figure 5.
Maximum likelihood phylogenetic trees of viruses detected with representative members that were previously identified in other mammalian and avian hosts. (a) Phylogenetic analysis of representative gammaretroviruses based on the 408 nucleotide (nt) sequence of a group-specific antigen (gag) gene. (b) Phylogenetic analysis of representative Bornaviridae based on partial RdRp amino acid sequences. (c) Phylogenetic analysis based on partial RdRp amino acid sequences of the novel paramyxovirus detected in this study with other previously identified representative members of Paramyxoviridae. The phylogenetic trees are midpoint rooted and the bar scales represent either the nucleotide or amino acid substitutions per site. Bootstrap values (70%) are shown at the key nodes. The new virus species detected in this study is shown in red in each tree. The silhouettes of figures were created with BioRender.com.