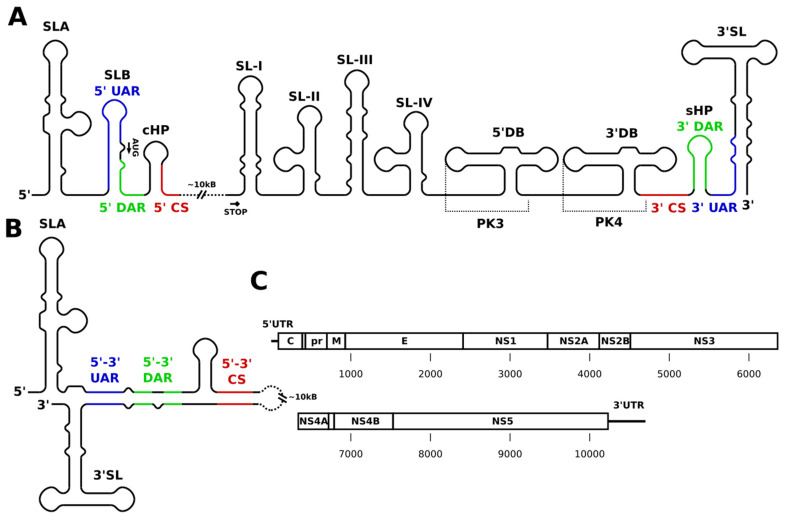
Figure 1.
Structural overview of flaviviral genomes. (A) Schematic representation of the linear 5′ and 3′ untranslated region (UTRs). Circularization motifs are indicated and labeled in color. The upstream AUG region (UAR; blue) is followed by the downstream AUG region (DAR; green) and the highly conserved circularization sequence (CS; red). Translation initiation happens within stem loop B (SLB) between 5′ UAR and 5′ DAR. The order of circularization sequence motifs is inverted in the 3′ region. (B) The circular form of the genome (paired motifs shown in color). SLB, capsid hairpin (cHP), short hairpin (sHP), and 3′SL undergo structural reorganization upon circularization. (C) Overview of flaviviral RNA genes. An ≈100 nt 5′ UTR is followed by a ≈10 kb single open reading frame coding a single genome polyprotein, which is post-translationally processed to form the structural (C, prM, and E) and non-structural proteins comprising the flaviviral proteome. The open reading frame is followed by an ≈300–700 nucleotides (depending on species) 3′ UTR containing conserved structural elements.