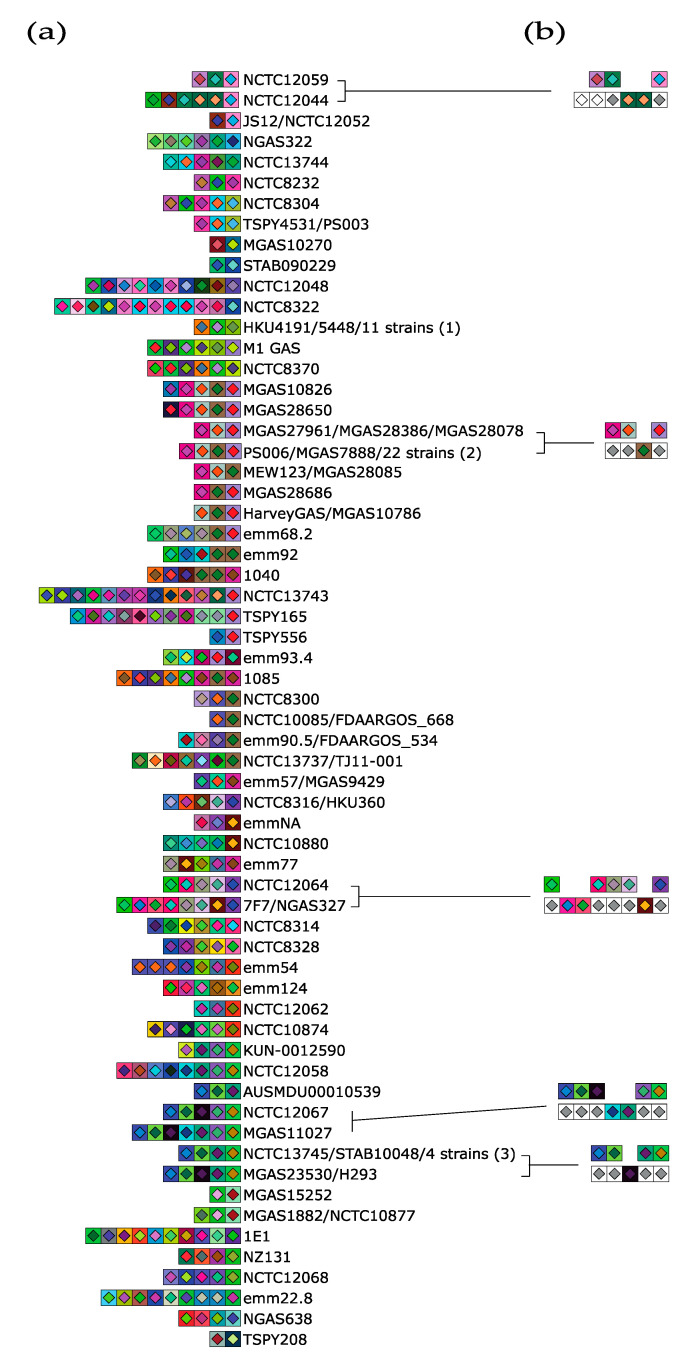
Figure 2.
Spacer representation of CRISPR type II-A loci of S. pyogenes genomes using CRISPRStudio. (a) Type II-A CRISPR loci from public S. pyogenes genomes and containing at least two spacers. Some strains have identical CRISPR arrays and it includes Group 1: Strains S119, MGAS2221, emm1, FDAARGOS_149, HKU488, 5448, 10-85, MTB314, M1 476, MGAS5005, A20. Group 2: MGAS7914, MGAS8347, MGAS11052 MGAS11108, MGAS11115, MGAS28191 MGAS28271, MGAS28278, MGAS28330, MGAS28360, MGAS28533, MGAS28669, MGAS28746, MGAS29064, MGAS29284, MGAS29326, MGAS29409, M28PF1, STAB09014, STAB10015, MGAS6180, NIH35. Group 3: STAB09023, MGAS27061, JMUB1235, KUN-0014944. (b) Examples of possible ectopic acquisition or insertion/deletion events in S. pyogenes CRISPR arrays. The order of the spacers is the same as in panel a. White square harboring a light gray diamond represents similar spacer. White square harboring a white diamond represents unique spacer on the 5′ end of the array.