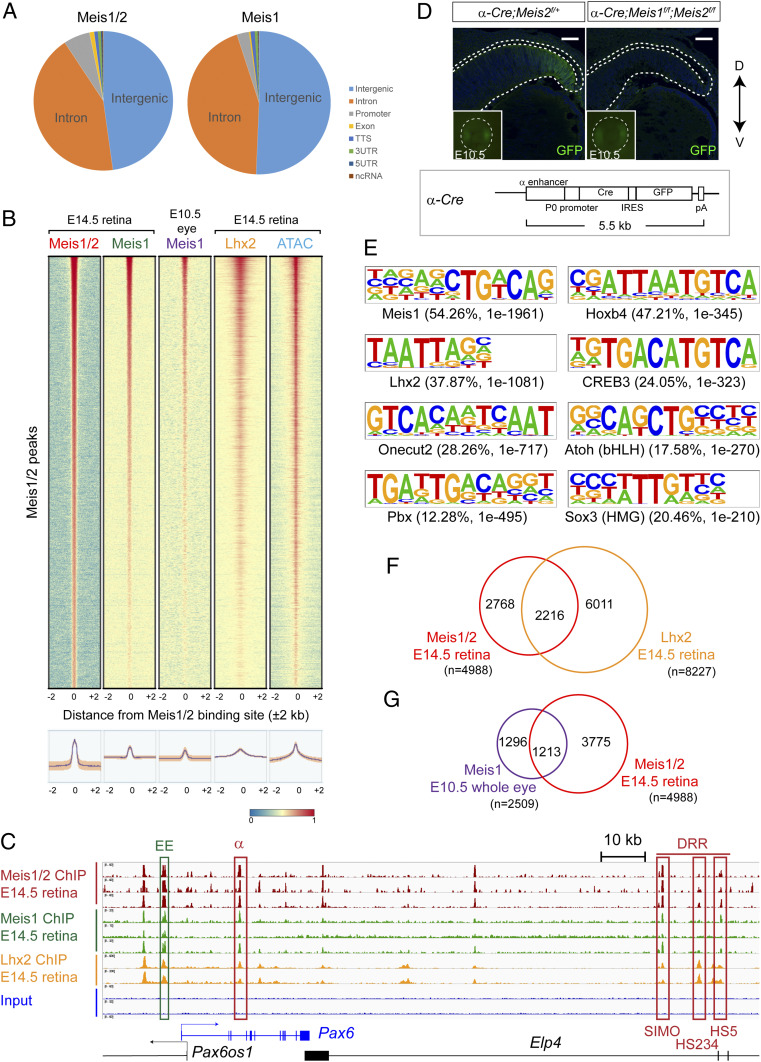
Fig. 2.
Meis1/2 directly regulate RPC-specific genes in concert with Lhx2. (A) Distribution of E14.5 retina Meis1/2 and Meis1 ChIP-seq peaks in the genome. (B) Density plots showing the distribution of Meis1 binding within the E10.5 whole eye [GSE62786 (27)], Meis1/2, Meis1 (E-MTAB-10112, present study), Lhx2 binding [GSE99818 (49)], and open chromatin region [GSE87064 (34)] in E14.5 neural retina at Meis1/2-bound regions (±2 kb from binding sites). The color scale shows the intensity of the distribution signal. The plotted lines were aligned according to E14.5 Meis1/2 ChIP-seq peaks. (C) ChIP-seq showing Meis1/2, Meis1, or Lhx2 binding in the E14.5 retina at Pax6. ChIP-seq peaks located within previously identified enhancers and promoters of Pax6 (downstream regulatory region [DRR] and α) are indicated by red boxes. (D) Frontal sections of E14.5 α-Cre;Meis2f/+ (control) and α-Cre;Meis1f/f;Meis2f/f embryos showing GFP signal. Insets show GFP signals from the heads of E10.5 embryos. The areas around the eye are shown in the Insets (Microscopic field: 400 × 400 μm). Schematic representation of α-Cre transgene. (Scale bar: 50 μm.) (E) DNA-binding motifs found in the Meis1/2-bound region by Homer. Percentage of target sequences with the motif and P value are indicated. (F) Venn diagram showing overlapping binding regions shared between Meis1/2 (E-MTAB-10112, present study) and Lhx2 [GSE99818 (48)] in E14.5 retina. (G) Venn diagram showing overlapping binding regions shared between Meis1/2 in E14.5 retina (E-MTAB-10112, present study) and Meis1 in E10.5 whole eye (27).